+Search query
-Structure paper
| Title | Binding of a negative allosteric modulator and competitive antagonist can occur simultaneously at the ionotropic glutamate receptor GluA2. |
|---|---|
| Journal, issue, pages | FEBS J, Vol. 288, Issue 3, Page 995-991007, Year 2021 |
| Publish date | Jul 8, 2020 |
 Authors Authors | Christian Krintel / Jerzy Dorosz / Andreas Haahr Larsen / Thor Seneca Thorsen / Raminta Venskutonytė / Osman Mirza / Michael Gajhede / Thomas Boesen / Jette Sandholm Kastrup /   |
| PubMed Abstract | Ionotropic glutamate receptors are ligand-gated ion channels governing neurotransmission in the central nervous system. Three major types of antagonists are known for the AMPA-type receptor GluA2: ...Ionotropic glutamate receptors are ligand-gated ion channels governing neurotransmission in the central nervous system. Three major types of antagonists are known for the AMPA-type receptor GluA2: competitive, noncompetitive (i.e., negative allosteric modulators; NAMs) used for treatment of epilepsy, and uncompetitive antagonists. We here report a 4.65 Å resolution X-ray structure of GluA2, revealing that four molecules of the competitive antagonist ZK200775 and four molecules of the NAM GYKI53655 are capable of binding at the same time. Using negative stain electron microscopy, we show that GYKI53655 alone or ZK200775/GYKI53655 in combination predominantly results in compact receptor forms. The agonist AMPA provides a mixed population of compact and bulgy shapes of GluA2 not impacted by addition of GYKI53655. Taken together, this suggests that the two different mechanisms of antagonism that lead to channel closure are independent and that the distribution between bulgy and compact receptors primarily depends on the ligand bound in the glutamate binding site. DATABASE: The atomic coordinates and structure factors from the crystal structure determination have been deposited in the Protein Data Bank under accession code https://doi.org/10.2210/pdb6RUQ/pdb. The electron microscopy 3D reconstruction volumes have been deposited in EMDB (EMD-4875: Apo; EMD-4920: ZK200775/GYKI53655; EMD-4921: AMPA compact; EMD-4922: AMPA/GYKI53655 bulgy; EMD-4923: GYKI53655; EMD-4924: AMPA bulgy; EMD-4925: AMPA/GYKI53655 compact). |
 External links External links |  FEBS J / FEBS J /  PubMed:32543078 PubMed:32543078 |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 4.65 - 26.9 Å |
| Structure data |  EMDB-4875:  EMDB-4920:  EMDB-4921:  EMDB-4922:  EMDB-4923:  EMDB-4924: 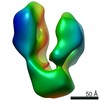 EMDB-4925:  PDB-6ruq: |
| Chemicals | 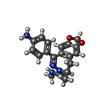 ChemComp-GYK: 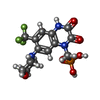 ChemComp-ZK1: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Receptor Negative allosteric modulator Antagonist Non-competitve antagonist |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




