+Search query
-Structure paper
| Title | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2. |
|---|---|
| Journal, issue, pages | Nat Microbiol, Vol. 4, Issue 3, Page 414-419, Year 2019 |
| Publish date | Dec 10, 2018 |
 Authors Authors | Daming Zhou / Yuguang Zhao / Abhay Kotecha / Elizabeth E Fry / James T Kelly / Xiangxi Wang / Zihe Rao / David J Rowlands / Jingshan Ren / David I Stuart /    |
| PubMed Abstract | Enterovirus 71 (EV71) is a common cause of hand, foot and mouth disease-a disease endemic especially in the Asia-Pacific region. Scavenger receptor class B member 2 (SCARB2) is the major receptor of ...Enterovirus 71 (EV71) is a common cause of hand, foot and mouth disease-a disease endemic especially in the Asia-Pacific region. Scavenger receptor class B member 2 (SCARB2) is the major receptor of EV71, as well as several other enteroviruses responsible for hand, foot and mouth disease, and plays a key role in cell entry. The isolated structures of EV71 and SCARB2 are known, but how they interact to initiate infection is not. Here, we report the EV71-SCARB2 complex structure determined at 3.4 Å resolution using cryo-electron microscopy. This reveals that SCARB2 binds EV71 on the southern rim of the canyon, rather than across the canyon, as predicted. Helices 152-163 (α5) and 183-193 (α7) of SCARB2 and the viral protein 1 (VP1) GH and VP2 EF loops of EV71 dominate the interaction, suggesting an allosteric mechanism by which receptor binding might facilitate the low-pH uncoating of the virus in the endosome/lysosome. Remarkably, many residues within the binding footprint are not conserved across SCARB2-dependent enteroviruses; however, a conserved proline and glycine seem to be key residues. Thus, although the virus maintains antigenic variability even within the receptor-binding footprint, the identification of binding 'hot spots' may facilitate the design of receptor mimic therapeutics less likely to quickly generate resistance. |
 External links External links |  Nat Microbiol / Nat Microbiol /  PubMed:30531980 PubMed:30531980 |
| Methods | EM (single particle) |
| Resolution | 3.4 Å |
| Structure data | |
| Chemicals | 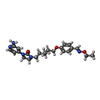 ChemComp-906:  ChemComp-NAG: |
| Source |
|
 Keywords Keywords | VIRUS / Enterovirus 71 / SCARB2 / Hand-foot-mouth disease / EV71 receptor |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 homo sapiens (human)
homo sapiens (human)
 enterovirus a71
enterovirus a71