+Search query
-Structure paper
| Title | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors. |
|---|---|
| Journal, issue, pages | Neuron, Vol. 99, Issue 5, Page 956-968.e4, Year 2018 |
| Publish date | Sep 5, 2018 |
 Authors Authors | Edward C Twomey / Maria V Yelshanskaya / Alexander A Vassilevski / Alexander I Sobolevsky /   |
| PubMed Abstract | AMPA receptors mediate fast excitatory neurotransmission and are critical for CNS development and function. Calcium-permeable subsets of AMPA receptors are strongly implicated in acute and chronic ...AMPA receptors mediate fast excitatory neurotransmission and are critical for CNS development and function. Calcium-permeable subsets of AMPA receptors are strongly implicated in acute and chronic neurological disorders. However, despite the clinical importance, the therapeutic landscape for specifically targeting them, and not the calcium-impermeable AMPA receptors, remains largely undeveloped. To address this problem, we used cryo-electron microscopy and electrophysiology to investigate the mechanisms by which small-molecule blockers selectively inhibit ion channel conductance in calcium-permeable AMPA receptors. We determined the structures of calcium-permeable GluA2 AMPA receptor complexes with the auxiliary subunit stargazin bound to channel blockers, including the orb weaver spider toxin AgTx-636, the spider toxin analog NASPM, and the adamantane derivative IEM-1460. Our structures provide insights into the architecture of the blocker binding site and the mechanism of trapping, which are critical for development of small molecules that specifically target calcium-permeable AMPA receptors. |
 External links External links |  Neuron / Neuron /  PubMed:30122377 / PubMed:30122377 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.9 - 4.8 Å |
| Structure data | EMDB-7959, PDB-6dlz: EMDB-7960, PDB-6dm0: |
| Chemicals |  ChemComp-GLU: 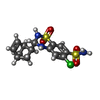 ChemComp-CYZ: 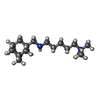 ChemComp-GZD: 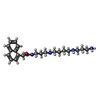 ChemComp-GYY:  ChemComp-LU7: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / Ion channel |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers












 homo sapiens (human)
homo sapiens (human)