Title Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase. Journal, issue, pages Eur. J. Med. Chem. , Vol. 234, Page 114270-114270, Year 2022Publish date Dec 1, 2021 (structure data deposition date) Nain-Perez, A. / Foller Fuchtbauer, A. / Haversen, L. / Lulla, A. / Gao, C. / Matic, J. / Monjas, L. / Rodriguez, A. / Brear, P. / Kim, W. ...Nain-Perez, A. / Foller Fuchtbauer, A. / Haversen, L. / Lulla, A. / Gao, C. / Matic, J. / Monjas, L. / Rodriguez, A. / Brear, P. / Kim, W. / Hyvonen, M. / Boren, J. / Mardinoglu, A. / Uhlen, M. / Grotli, M. / Methods X-ray diffraction Resolution 1.685 - 2.354 Å Structure data PDB-5sc8 17 Method : X-RAY DIFFRACTION / Resolution : 1.77 Å
PDB-5sc9 29 Method : X-RAY DIFFRACTION / Resolution : 1.685 Å
PDB-5sca 36 Method : X-RAY DIFFRACTION / Resolution : 1.918 Å
PDB-5scb 28 Method : X-RAY DIFFRACTION / Resolution : 1.8 Å
PDB-5scc 57 Method : X-RAY DIFFRACTION / Resolution : 1.885 Å
PDB-5scd 58 Method : X-RAY DIFFRACTION / Resolution : 2.041 Å
PDB-5sce 55 Method : X-RAY DIFFRACTION / Resolution : 2.147 Å
PDB-5scf 99 Method : X-RAY DIFFRACTION / Resolution : 2.185 Å
PDB-5scg 101 Method : X-RAY DIFFRACTION / Resolution : 1.937 Å
PDB-5sch 100 Method : X-RAY DIFFRACTION / Resolution : 2.089 Å
PDB-5sci 105 Method : X-RAY DIFFRACTION / Resolution : 2.155 Å
PDB-5scj 106 Method : X-RAY DIFFRACTION / Resolution : 2.354 Å
PDB-5sck 42 Method : X-RAY DIFFRACTION / Resolution : 1.717 Å
PDB-5scl 1 Method : X-RAY DIFFRACTION / Resolution : 2.134 Å
PDB-5sdt 15 Method : X-RAY DIFFRACTION / Resolution : 1.944 Å
PDB-7qdn human liver pyruvate kinase from which the B domain has been deleted Method : X-RAY DIFFRACTION / Resolution : 1.695 Å
PDB-7qzu 47 Method : X-RAY DIFFRACTION / Resolution : 1.964 Å
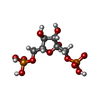
Chemicals ChemComp-FBP
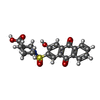
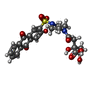
ChemComp-E6K
ChemComp-I7K
ChemComp-I7N
ChemComp-I7V
ChemComp-I8Q
ChemComp-I8U
ChemComp-I8Y
ChemComp-I91
ChemComp-I99
ChemComp-I9C
ChemComp-I9F
ChemComp-I9K
ChemComp-I9N
ChemComp-I9Q
ChemComp-I4L
Source homo sapiens (human) / / / / / / /
 Authors
Authors External links
External links Eur. J. Med. Chem. /
Eur. J. Med. Chem. /  PubMed:35290845
PubMed:35290845

































 Keywords
Keywords Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)