+Search query
-Structure paper
| Title | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library |
|---|---|
| Journal, issue, pages | To Be Published |
| Publish date | Mar 12, 2019 (structure data deposition date) |
 Authors Authors | Petrick, J.K. / Muenzker, L. / von Delft, F. / Jahnke, W. |
 External links External links | Search PubMed |
| Methods | X-ray diffraction |
| Resolution | 1.41 - 2.2 Å |
| Structure data |  PDB-5qpd:  PDB-5qpe:  PDB-5qpf:  PDB-5qpg:  PDB-5qph:  PDB-5qpi:  PDB-5qpj:  PDB-5qpk:  PDB-5qpl:  PDB-5qpm:  PDB-5qpn:  PDB-5qpo:  PDB-5qpp:  PDB-5qpq:  PDB-5qpr:  PDB-5qps:  PDB-5qpt:  PDB-5qpu:  PDB-5qpv:  PDB-5qpw:  PDB-5qpx:  PDB-5qpy:  PDB-5qpz:  PDB-5qq0:  PDB-5qq1:  PDB-5qq2:  PDB-5qq3:  PDB-5qq4:  PDB-5qq5:  PDB-5qq6:  PDB-5qq7:  PDB-5qq8:  PDB-5qq9:  PDB-5qqa:  PDB-5qqb: |
| Chemicals |  ChemComp-SO4:  ChemComp-ACT:  ChemComp-ZN:  ChemComp-LT7:  ChemComp-HOH:  ChemComp-AWG:  ChemComp-AWM:  ChemComp-AWV:  ChemComp-LUS:  ChemComp-GQM: 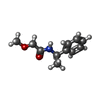 ChemComp-JGJ:  ChemComp-LUY:  ChemComp-M0J: 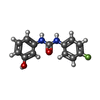 ChemComp-LV1:  ChemComp-LDV:  ChemComp-GQP:  ChemComp-LV4: 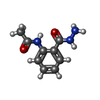 ChemComp-LV7: 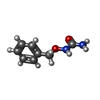 ChemComp-LVD:  ChemComp-LVP:  ChemComp-LVV:  ChemComp-JHS:  ChemComp-LWA:  ChemComp-JH7:  ChemComp-LWD:  ChemComp-JH1:  ChemComp-AYV:  ChemComp-LWV:  ChemComp-LX4:  ChemComp-MJ4:  ChemComp-LXA:  ChemComp-LX7:  ChemComp-JJM:  ChemComp-LXJ:  ChemComp-LXM:  ChemComp-LXS:  ChemComp-M0D:  ChemComp-LZV:  ChemComp-LZY: |
| Source |
|
 Keywords Keywords | TRANSFERASE / SGC - Diamond I04-1 fragment screening / PanDDA / XChemExplorer |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




