+Search query
-Structure paper
| Title | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors. |
|---|---|
| Journal, issue, pages | J. Med. Chem., Vol. 53, Page 942-950, Year 2010 |
| Publish date | Nov 11, 2009 (structure data deposition date) |
 Authors Authors | Wang, Y.S. / Strickland, C. / Voigt, J.H. / Kennedy, M.E. / Beyer, B.M. / Senior, M.M. / Smith, E.M. / Nechuta, T.L. / Madison, V.S. / Czarniecki, M. ...Wang, Y.S. / Strickland, C. / Voigt, J.H. / Kennedy, M.E. / Beyer, B.M. / Senior, M.M. / Smith, E.M. / Nechuta, T.L. / Madison, V.S. / Czarniecki, M. / McKittrick, B.A. / Stamford, A.W. / Parker, E.M. / Hunter, J.C. / Greenlee, W.J. / Wyss, D.F. |
 External links External links |  J. Med. Chem. / J. Med. Chem. /  PubMed:20043700 PubMed:20043700 |
| Methods | X-ray diffraction |
| Resolution | 1.7 - 1.9 Å |
| Structure data |  PDB-3kmx:  PDB-3kmy:  PDB-3kn0: |
| Chemicals | 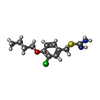 ChemComp-G00:  ChemComp-HOH: 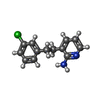 ChemComp-D8Y: 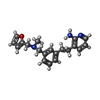 ChemComp-3TO:  ChemComp-TLA: |
| Source |
|
 Keywords Keywords | HYDROLASE / Alzheimer's / BACE1 / Alternative splicing / Aspartyl protease / Disulfide bond / Endoplasmic reticulum / Endosome / Glycoprotein / Golgi apparatus / Membrane / Polymorphism / Protease / Transmembrane / Zymogen |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)