+Search query
-Structure paper
| Title | Fragment-based discovery of hepatitis C virus NS5b RNA polymerase inhibitors. |
|---|---|
| Journal, issue, pages | Bioorg. Med. Chem. Lett., Vol. 18, Page 2990-2995, Year 2008 |
| Publish date | Mar 12, 2008 (structure data deposition date) |
 Authors Authors | Antonysamy, S.S. / Aubol, B. / Blaney, J. / Browner, M.F. / Giannetti, A.M. / Harris, S.F. / Hebert, N. / Hendle, J. / Hopkins, S. / Jefferson, E. ...Antonysamy, S.S. / Aubol, B. / Blaney, J. / Browner, M.F. / Giannetti, A.M. / Harris, S.F. / Hebert, N. / Hendle, J. / Hopkins, S. / Jefferson, E. / Kissinger, C. / Leveque, V. / Marciano, D. / McGee, E. / Najera, I. / Nolan, B. / Tomimoto, M. / Torres, E. / Wright, T. |
 External links External links |  Bioorg. Med. Chem. Lett. / Bioorg. Med. Chem. Lett. /  PubMed:18400495 PubMed:18400495 |
| Methods | X-ray diffraction |
| Resolution | 1.75 - 2.07 Å |
| Structure data |  PDB-3ciz:  PDB-3cj0:  PDB-3cj2:  PDB-3cj3:  PDB-3cj4:  PDB-3cj5: |
| Chemicals |  ChemComp-ZN: 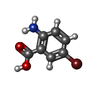 ChemComp-SX1:  ChemComp-HOH:  ChemComp-SX2:  ChemComp-NI: 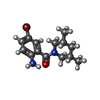 ChemComp-SX3: 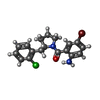 ChemComp-SX4: 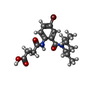 ChemComp-SX5: 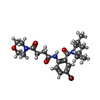 ChemComp-SX6: |
| Source |
|
 Keywords Keywords | TRANSFERASE / DRUG DISCOVERY / HCV NS5B / FRAGMENT BASED / Apoptosis / ATP-binding / Capsid protein / Endoplasmic reticulum / Envelope protein / Fusion protein / Glycoprotein / Helicase / Host-virus interaction / Hydrolase / Interferon antiviral system evasion / Lipid droplet / Lipoprotein / Membrane / Metal-binding / Mitochondrion / Multifunctional enzyme / Nucleotide-binding / Nucleotidyltransferase / Nucleus / Oncogene / Palmitate / Phosphoprotein / Protease / Ribonucleoprotein / RNA replication / RNA-binding / RNA-directed RNA polymerase / Secreted / Serine protease / SH3-binding / Thiol protease / Transcription / Transcription regulation / Transmembrane / Viral nucleoprotein / Virion / Acetylation / Cytoplasm / Ubl conjugation / Zinc |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 hepatitis c virus subtype 1b
hepatitis c virus subtype 1b