+検索条件
-Structure paper
| タイトル | Structural mechanism for statin inhibition of HMG-CoA reductase. |
|---|---|
| ジャーナル・号・ページ | Science, Vol. 292, Page 1160-1164, Year 2001 |
| 掲載日 | 2001年1月9日 (構造データの登録日) |
 著者 著者 | E S Istvan / J Deisenhofer /  |
| PubMed 要旨 | HMG-CoA (3-hydroxy-3-methylglutaryl-coenzyme A) reductase (HMGR) catalyzes the committed step in cholesterol biosynthesis. Statins are HMGR inhibitors with inhibition constant values in the nanomolar ...HMG-CoA (3-hydroxy-3-methylglutaryl-coenzyme A) reductase (HMGR) catalyzes the committed step in cholesterol biosynthesis. Statins are HMGR inhibitors with inhibition constant values in the nanomolar range that effectively lower serum cholesterol levels and are widely prescribed in the treatment of hypercholesterolemia. We have determined structures of the catalytic portion of human HMGR complexed with six different statins. The statins occupy a portion of the binding site of HMG-CoA, thus blocking access of this substrate to the active site. Near the carboxyl terminus of HMGR, several catalytically relevant residues are disordered in the enzyme-statin complexes. If these residues were not flexible, they would sterically hinder statin binding. |
 リンク リンク |  Science / Science /  PubMed:11349148 PubMed:11349148 |
| 手法 | X線回折 |
| 解像度 | 2.1 - 2.33 Å |
| 構造データ |  PDB-1hw8:  PDB-1hw9:  PDB-1hwi:  PDB-1hwj:  PDB-1hwk:  PDB-1hwl: |
| 化合物 | 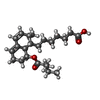 ChemComp-114:  ChemComp-ADP:  ChemComp-HOH:  ChemComp-SIM:  ChemComp-115: 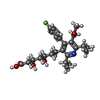 ChemComp-116: 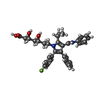 ChemComp-117:  ChemComp-FBI: |
| 由来 |
|
 キーワード キーワード | OXIDOREDUCTASE / protein-inhibitor complex |
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア 万見文献について
万見文献について



 homo sapiens (ヒト)
homo sapiens (ヒト)