+Search query
-Structure paper
| Title | A single allosteric site merges activation, modulation and inhibition in TRPM5. |
|---|---|
| Journal, issue, pages | Nat Chem Biol, Year 2026 |
| Publish date | Jan 5, 2026 |
 Authors Authors | Zheng Ruan / Junuk Lee / Yangyang Li / Ian J Orozco / Juan Du / Wei Lü /  |
| PubMed Abstract | TRPM5 is a Ca-activated monovalent cation channel essential for taste perception, insulin secretion and gastrointestinal chemosensation. Canonical TRPM5 activation requires Ca binding at two distinct ...TRPM5 is a Ca-activated monovalent cation channel essential for taste perception, insulin secretion and gastrointestinal chemosensation. Canonical TRPM5 activation requires Ca binding at two distinct sites: an agonist site within the lower vestibule of the S1-S4 pocket in the transmembrane domain (Ca) and a modulatory site in the intracellular domain (Ca) that tunes voltage dependence and agonist sensitivity. Here we characterize CBTA as a noncalcium agonist that binds to the upper vestibule of the S1-S4 pocket, directly above Ca. CBTA alone mimics the dual role of Ca and Ca, merging agonist activation with voltage modulation. CBTA also renders TRPM5 supersensitive to Ca, synergistically hyperactivating the channel even at near-resting Ca levels. We further demonstrate that the inhibitor triphenylphosphine oxide binds the same site but stabilizes a nonconductive state. These opposing effects reveal the upper S1-S4 pocket as a multifunctional regulatory hub integrating activation, inhibition and modulation in TRPM5. |
 External links External links |  Nat Chem Biol / Nat Chem Biol /  PubMed:41491833 PubMed:41491833 |
| Methods | EM (single particle) |
| Resolution | 2.75 - 3.5 Å |
| Structure data |  EMDB-71244: The consensus map of zebrafish TRPM5 with 1mM EDTA and 0.5mM CBTA EMDB-71245, PDB-9p3n: EMDB-71246, PDB-9p3o:  EMDB-71247: Zebrafish TRPM5 with 0.2mM calcium  EMDB-71248: Zebrafish TRPM5 with 1uM calcium  EMDB-71249: Zebrafish TRPM5 with 2uM Ca in GDN detergent EMDB-71250, PDB-9p3p: EMDB-71251, PDB-9p3q: EMDB-71252, PDB-9p3r: EMDB-71253, PDB-9p3s: EMDB-71254, PDB-9p3t: EMDB-71255, PDB-9p3u: EMDB-71256, PDB-9p3v: EMDB-71257, PDB-9p3w: |
| Chemicals |  ChemComp-CA:  ChemComp-YUY: 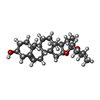 ChemComp-YUV:  PDB-1cgw:  ChemComp-NAG:  PDB-1cgz:  PDB-1cg0:  PDB-1cg5:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / TRPM5 channel; ion channel; cation channel; sodium channel; calcium; CBTA / TRPM5 channel / ion channel / cation channel / sodium channel / TRANSFERASE |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers
























