+Search query
-Structure paper
| Title | Deciphering the assembly process of PQQ dependent methanol dehydrogenase. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 16, Issue 1, Page 6672, Year 2025 |
| Publish date | Jul 19, 2025 |
 Authors Authors | Haichuan Zhou / Junqing Sun / Jian Cheng / Min Wu / Jie Bai / Qian Li / Jie Shen / Manman Han / Chen Yang / Liangpo Li / Yuwan Liu / Qichen Cao / Weidong Liu / Haixia Xiao / Hongjun Dong / Feng Gao / Huifeng Jiang /  |
| PubMed Abstract | Pyrroloquinoline quinone (PQQ)-dependent methanol dehydrogenases (MDHs), the periplasmic metalloenzymes in Gram-negative methylotrophic bacteria, play a pivotal role in methane and methanol bio- ...Pyrroloquinoline quinone (PQQ)-dependent methanol dehydrogenases (MDHs), the periplasmic metalloenzymes in Gram-negative methylotrophic bacteria, play a pivotal role in methane and methanol bio-utilization. Although the structures of many PQQ-dependent MDHs have been resolved, including the canonical heterotetrameric enzymes composed of two MxaF and two MxaI subunits with a molecule of PQQ and a calcium ion in the active site in MxaF, the biogenesis of these enzymes remains elusive. Here, we characterize a chaperone, MxaJ, responsible for PQQ incorporation by reconstructing a PQQ-dependent MDH assembly system in Escherichia coli. Using cryo-electron microscopy, we capture the structures of the intermediate complexes formed by the chaperone MxaJ and catalytic subunit MxaF during PQQ-dependent MDH maturation, revealing a chaperone-mediated molecular mechanism of cofactor incorporation. These findings not only advance our understanding on the biogenesis of PQQ-dependent MDH, but also provide an alternative engineering way for methane and methanol bioconversion. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:40683858 / PubMed:40683858 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.78 - 2.81 Å |
| Structure data | EMDB-60838, PDB-9ism: EMDB-60840, PDB-9iso: |
| Chemicals | 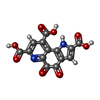 ChemComp-PQQ: |
| Source |
|
 Keywords Keywords | PROTEIN BINDING / MxaF/MxaJ / MxaF/MxaJ/PQQ |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 methylorubrum extorquens (bacteria)
methylorubrum extorquens (bacteria)