+Search query
-Structure paper
| Title | Distinct binding conformations of epinephrine with α- and β-adrenergic receptors. |
|---|---|
| Journal, issue, pages | Exp Mol Med, Vol. 56, Issue 9, Page 1952-1966, Year 2024 |
| Publish date | Sep 2, 2024 |
 Authors Authors | Jian-Shu Lou / Minfei Su / Jinan Wang / Hung Nguyen Do / Yinglong Miao / Xin-Yun Huang /   |
| PubMed Abstract | Agonists targeting α-adrenergic receptors (ARs) are used to treat diverse conditions, including hypertension, attention-deficit/hyperactivity disorder, pain, panic disorders, opioid and alcohol ...Agonists targeting α-adrenergic receptors (ARs) are used to treat diverse conditions, including hypertension, attention-deficit/hyperactivity disorder, pain, panic disorders, opioid and alcohol withdrawal symptoms, and cigarette cravings. These receptors transduce signals through heterotrimeric Gi proteins. Here, we elucidated cryo-EM structures that depict α-AR in complex with Gi proteins, along with the endogenous agonist epinephrine or the synthetic agonist dexmedetomidine. Molecular dynamics simulations and functional studies reinforce the results of the structural revelations. Our investigation revealed that epinephrine exhibits different conformations when engaging with α-ARs and β-ARs. Furthermore, α-AR and β-AR (primarily coupled to Gs, with secondary associations to Gi) were compared and found to exhibit different interactions with Gi proteins. Notably, the stability of the epinephrine-α-AR-Gi complex is greater than that of the dexmedetomidine-α-AR-Gi complex. These findings substantiate and improve our knowledge on the intricate signaling mechanisms orchestrated by ARs and concurrently shed light on the regulation of α-ARs and β-ARs by epinephrine. |
 External links External links |  Exp Mol Med / Exp Mol Med /  PubMed:39218975 / PubMed:39218975 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.8 - 3.2 Å |
| Structure data | EMDB-45425, PDB-9cbl: EMDB-45426, PDB-9cbm: |
| Chemicals | 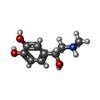 ChemComp-ALE: 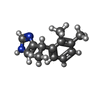 ChemComp-CZX: |
| Source |
|
 Keywords Keywords | SIGNALING PROTEIN / GPCR / Adrenergic Receptor / GPCR Adrenergic Receptor |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 homo sapiens (human)
homo sapiens (human)
 enterobacteria phage t4 (virus)
enterobacteria phage t4 (virus)