+Search query
-Structure paper
| Title | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 10226, Year 2024 |
| Publish date | Nov 26, 2024 |
 Authors Authors | Shen-Ming Huang / Zhi-Zhen Huang / Lei Liu / Meng-Yao Xiong / Chao Zhang / Bo-Yang Cai / Ming-Wei Wang / Kui Cai / Ying-Li Jia / Jia-Le Wang / Ming-Hui Zhang / Yi-He Xie / Min Li / Hang Zhang / Cheng-Hao Weng / Xin Wen / Zhi Li / Ying Sun / Fan Yi / Zhao Yang / Peng Xiao / Fan Yang / Xiao Yu / Lu Tie / Bao-Xue Yang / Jin-Peng Sun /  |
| PubMed Abstract | Urea's transmembrane transport through urea transporters (UT) is a fundamental physiological behavior for life activities. Here, we present 11 cryo-EM structures of four UT members in resting states, ...Urea's transmembrane transport through urea transporters (UT) is a fundamental physiological behavior for life activities. Here, we present 11 cryo-EM structures of four UT members in resting states, urea transport states, or inactive states bound with synthetic competitive, uncompetitive or noncompetitive inhibitor. Our results indicate that the binding of urea via a conserved urea recognition motif (URM) and the urea transport via H-bond transfer along the Q-T-T-Q motif among different UT members. Moreover, distinct binding modes of the competitive inhibitors 25a and ATB3, the uncompetitive inhibitor CF11 and the noncompetitive inhibitor HQA2 provide different mechanisms for blocking urea transport and achieved selectivity through L-P pocket, UCBP region and SCG pocket, respectively. In summary, our study not only allows structural understanding of urea transport via UTs but also afforded a structural landscape of hUT-A2 inhibition by competitive, uncompetitive and noncompetitive inhibitors, which may facilitate developing selective human UT-A inhibitors as a new class of salt-sparing diuretics. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:39587082 / PubMed:39587082 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.3 - 3.3 Å |
| Structure data | EMDB-38268, PDB-8xd7: EMDB-38270, PDB-8xd9: EMDB-38271, PDB-8xda: EMDB-38272, PDB-8xdb: EMDB-38273, PDB-8xdc: EMDB-38274, PDB-8xdd: EMDB-38275, PDB-8xde: EMDB-38276, PDB-8xdf: EMDB-38277, PDB-8xdg: EMDB-38278, PDB-8xdh: EMDB-38279, PDB-8xdi: |
| Chemicals |  PDB-1lu9: 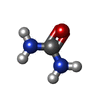 ChemComp-URE:  ChemComp-HOH:  PDB-1lyj:  PDB-1lva: 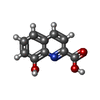 ChemComp-8HC: 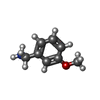 ChemComp-SZ4: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Urea transporter |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers

























 homo sapiens (human)
homo sapiens (human)
