+Search query
-Structure paper
| Title | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket. |
|---|---|
| Journal, issue, pages | EMBO J, Vol. 42, Issue 17, Page e113415, Year 2023 |
| Publish date | Sep 4, 2023 |
 Authors Authors | Jie Wang / Xu Li / Fang-Fang Wang / Meng-Ting Cheng / Yao-Xu Mao / Shu-Cheng Fang / Liang Wang / Cong-Zhao Zhou / Wen-Tao Hou / Yuxing Chen /  |
| PubMed Abstract | The human ABC transporter ABCC3 (also known as MRP3) transports a wide spectrum of substrates, including endogenous metabolites and exogenous drugs. Accordingly, it participates in multiple ...The human ABC transporter ABCC3 (also known as MRP3) transports a wide spectrum of substrates, including endogenous metabolites and exogenous drugs. Accordingly, it participates in multiple physiological processes and is involved in diverse human diseases such as intrahepatic cholestasis of pregnancy, which is caused by the intracellular accumulation of bile acids and estrogens. Here, we report three cryogenic electron microscopy structures of ABCC3: in the apo-form and in complexed forms bound to either the conjugated sex hormones β-estradiol 17-(β-D-glucuronide) and dehydroepiandrosterone sulfate. For both hormones, the steroid nuclei that superimpose against each other occupy the hydrophobic center of the transport cavity, whereas the two conjugation groups are separated and fixed by the hydrophilic patches in two transmembrane domains. Structural analysis combined with site-directed mutagenesis and ATPase activity assays revealed that ABCC3 possesses an amphiphilic substrate-binding pocket able to hold either conjugated hormone in an asymmetric pattern. These data build on consensus features of the substrate-binding pocket of MRPs and provide a structural platform for the rational design of inhibitors. |
 External links External links |  EMBO J / EMBO J /  PubMed:37485728 / PubMed:37485728 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.07 - 3.65 Å |
| Structure data | EMDB-35043, PDB-8hvh: EMDB-35049, PDB-8hw2: EMDB-35050, PDB-8hw4: |
| Chemicals | 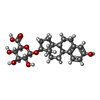 ChemComp-U38: 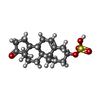 ChemComp-ZWY: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / ABC transporter / TRANSPORT PROTEIN / multidrug resistance protein |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 homo sapiens (human)
homo sapiens (human)