+検索条件
-Structure paper
| タイトル | Deep learning driven de novo drug design based on gastric proton pump structures. |
|---|---|
| ジャーナル・号・ページ | Commun Biol, Vol. 6, Issue 1, Page 956, Year 2023 |
| 掲載日 | 2023年9月19日 |
 著者 著者 | Kazuhiro Abe / Mami Ozako / Miki Inukai / Yoe Matsuyuki / Shinnosuke Kitayama / Chisato Kanai / Chiaki Nagai / Chai C Gopalasingam / Christoph Gerle / Hideki Shigematsu / Nariyoshi Umekubo / Satoshi Yokoshima / Atsushi Yoshimori /  |
| PubMed 要旨 | Existing drugs often suffer in their effectiveness due to detrimental side effects, low binding affinity or pharmacokinetic problems. This may be overcome by the development of distinct compounds. ...Existing drugs often suffer in their effectiveness due to detrimental side effects, low binding affinity or pharmacokinetic problems. This may be overcome by the development of distinct compounds. Here, we exploit the rich structural basis of drug-bound gastric proton pump to develop compounds with strong inhibitory potency, employing a combinatorial approach utilizing deep generative models for de novo drug design with organic synthesis and cryo-EM structural analysis. Candidate compounds that satisfy pharmacophores defined in the drug-bound proton pump structures, were designed in silico utilizing our deep generative models, a workflow termed Deep Quartet. Several candidates were synthesized and screened according to their inhibition potencies in vitro, and their binding poses were in turn identified by cryo-EM. Structures reaching up to 2.10 Å resolution allowed us to evaluate and re-design compound structures, heralding the most potent compound in this study, DQ-18 (N-methyl-4-((2-(benzyloxy)-5-chlorobenzyl)oxy)benzylamine), which shows a K value of 47.6 nM. Further high-resolution cryo-EM analysis at 2.08 Å resolution unambiguously determined the DQ-18 binding pose. Our integrated approach offers a framework for structure-based de novo drug development based on the desired pharmacophores within the protein structure. |
 リンク リンク |  Commun Biol / Commun Biol /  PubMed:37726448 / PubMed:37726448 /  PubMed Central PubMed Central |
| 手法 | EM (単粒子) |
| 解像度 | 2.08 - 2.26 Å |
| 構造データ | EMDB-35500, PDB-8ijv: EMDB-35501, PDB-8ijw: EMDB-35502, PDB-8ijx: EMDB-36424, PDB-8jmn: |
| 化合物 |  ChemComp-PCW:  ChemComp-MG: 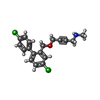 ChemComp-PWI:  ChemComp-NAG:  ChemComp-CLR:  ChemComp-HOH: 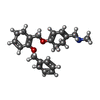 ChemComp-PXR: 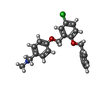 ChemComp-PZ0: 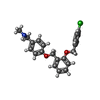 ChemComp-UOU: |
| 由来 |
|
 キーワード キーワード | MEMBRANE PROTEIN / P-type ATPase / P2-type ATPase / proton pump / gastric |
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア 万見文献について
万見文献について












