+Search query
-Structure paper
| Title | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes. |
|---|---|
| Journal, issue, pages | Cell, Vol. 184, Issue 4, Page 943-956.e18, Year 2021 |
| Publish date | Feb 18, 2021 |
 Authors Authors | Peng Xiao / Wei Yan / Lu Gou / Ya-Ni Zhong / Liangliang Kong / Chao Wu / Xin Wen / Yuan Yuan / Sheng Cao / Changxiu Qu / Xin Yang / Chuan-Cheng Yang / Anjie Xia / Zhenquan Hu / Qianqian Zhang / Yong-Hao He / Dao-Lai Zhang / Chao Zhang / Gui-Hua Hou / Huanxiang Liu / Lizhe Zhu / Ping Fu / Shengyong Yang / Daniel M Rosenbaum / Jin-Peng Sun / Yang Du / Lei Zhang / Xiao Yu / Zhenhua Shao /   |
| PubMed Abstract | Dopamine receptors, including D1- and D2-like receptors, are important therapeutic targets in a variety of neurological syndromes, as well as cardiovascular and kidney diseases. Here, we present five ...Dopamine receptors, including D1- and D2-like receptors, are important therapeutic targets in a variety of neurological syndromes, as well as cardiovascular and kidney diseases. Here, we present five cryoelectron microscopy (cryo-EM) structures of the dopamine D1 receptor (DRD1) coupled to Gs heterotrimer in complex with three catechol-based agonists, a non-catechol agonist, and a positive allosteric modulator for endogenous dopamine. These structures revealed that a polar interaction network is essential for catecholamine-like agonist recognition, whereas specific motifs in the extended binding pocket were responsible for discriminating D1- from D2-like receptors. Moreover, allosteric binding at a distinct inner surface pocket improved the activity of DRD1 by stabilizing endogenous dopamine interaction at the orthosteric site. DRD1-Gs interface revealed key features that serve as determinants for G protein coupling. Together, our study provides a structural understanding of the ligand recognition, allosteric regulation, and G protein coupling mechanisms of DRD1. |
 External links External links |  Cell / Cell /  PubMed:33571432 / PubMed:33571432 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.1 - 3.54 Å |
| Structure data | EMDB-30392, PDB-7ckw: EMDB-30393, PDB-7ckx: EMDB-30394, PDB-7cky: EMDB-30395, PDB-7ckz: EMDB-30452, PDB-7crh: |
| Chemicals |  ChemComp-G3C:  ChemComp-CLR:  ChemComp-G3O: 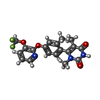 ChemComp-G3U:  ChemComp-LDP: 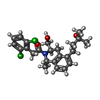 ChemComp-G4C:  ChemComp-GBU: |
| Source |
|
 Keywords Keywords |  CELL CYCLE / CELL CYCLE /  transmembrane protein / signaling transduction / transmembrane protein / signaling transduction /  transduction / transduction /  complex / complex /  signaling complex / signaling complex /  cell signaling cell signaling |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















