+Search query
-Structure paper
| Title | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening |
|---|---|
| Journal, issue, pages | To be Published |
| Publish date | Feb 2, 2011 (structure data deposition date) |
 Authors Authors | Schonbrunn, E. / Becker, A. / Betzi, S. / Alam, R. / Han, H. / Rawle, F. / Katta, V. / Jakkaraj, J. / Chakrasali, R. / Neelam, S. ...Schonbrunn, E. / Becker, A. / Betzi, S. / Alam, R. / Han, H. / Rawle, F. / Katta, V. / Jakkaraj, J. / Chakrasali, R. / Neelam, S. / Hook, D. / Tash, J. / Georg, G. |
 External links External links | Search PubMed |
| Methods | X-ray diffraction |
| Resolution | 1.6 - 2 Å |
| Structure data |  PDB-3ql8:  PDB-3qqf:  PDB-3qqg:  PDB-3qqh:  PDB-3qqj:  PDB-3qql:  PDB-3qrt:  PDB-3qru:  PDB-3qwj:  PDB-3qwk:  PDB-3qx2:  PDB-3qx4:  PDB-3qxo:  PDB-3qzf:  PDB-3qzg:  PDB-3qzh:  PDB-3qzi:  PDB-3r1q:  PDB-3r1s:  PDB-3r1y:  PDB-3r28:  PDB-3r6x:  PDB-3r71:  PDB-3r73:  PDB-3r7e:  PDB-3r7i:  PDB-3r7u:  PDB-3r7v:  PDB-3r7y:  PDB-3r83:  PDB-3r8l:  PDB-3r8m:  PDB-3r8p:  PDB-3rai:  PDB-3rm6:  PDB-3rm7:  PDB-3roy:  PDB-3rpo: |
| Chemicals |  ChemComp-EDO: 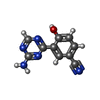 ChemComp-X01:  ChemComp-HOH:  ChemComp-X07:  ChemComp-X06:  ChemComp-PO4:  ChemComp-X0A:  ChemComp-X11:  ChemComp-X03: 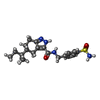 ChemComp-X14:  ChemComp-X19:  ChemComp-X6A:  ChemComp-X62:  ChemComp-X63:  ChemComp-X4B: 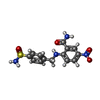 ChemComp-X65:  ChemComp-X66:  ChemComp-X67:  ChemComp-X69:  ChemComp-X72:  ChemComp-X75: 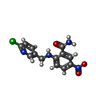 ChemComp-X73:  ChemComp-X76:  ChemComp-XA0:  ChemComp-X84:  ChemComp-X86:  ChemComp-X87:  ChemComp-X88:  ChemComp-X9I:  ChemComp-X96:  ChemComp-Z02:  ChemComp-Z04:  ChemComp-Z14:  ChemComp-Z30:  ChemComp-Z19:  ChemComp-Z46:  ChemComp-X85:  ChemComp-18Z:  ChemComp-19Z:  ChemComp-22Z:  ChemComp-24Z: |
| Source |
|
 Keywords Keywords | TRANSFERASE/TRANSFERASE INHIBITOR / protein kinase / inhibitor / TRANSFERASE-TRANSFERASE INHIBITOR complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)