+Search query
-Structure paper
| Title | Amyloidogenesis of bacterial prionoid RepA-WH1 recapitulates dimer to monomer transitions of RepA in DNA replication initiation. |
|---|---|
| Journal, issue, pages | Structure, Vol. 23, Issue 1, Page 183-189, Year 2015 |
| Publish date | Jan 6, 2015 |
 Authors Authors | Eva Torreira / María Moreno-Del Álamo / Maria Eugenia Fuentes-Perez / Cristina Fernández / Jaime Martín-Benito / Fernando Moreno-Herrero / Rafael Giraldo / Oscar Llorca /  |
| PubMed Abstract | Most available structures of amyloids correspond to peptide fragments that self-assemble in extended cross β sheets. However, structures in which a whole protein domain acts as building block of an ...Most available structures of amyloids correspond to peptide fragments that self-assemble in extended cross β sheets. However, structures in which a whole protein domain acts as building block of an amyloid fiber are scarce, in spite of their relevance to understand amyloidogenesis. Here, we use electron microscopy (EM) and atomic force microscopy (AFM) to analyze the structure of amyloid filaments assembled by RepA-WH1, a winged-helix domain from a DNA replication initiator in bacterial plasmids. RepA-WH1 functions as a cytotoxic bacterial prionoid that recapitulates features of mammalian amyloid proteinopathies. RepA are dimers that monomerize at the origin to initiate replication, and we find that RepA-WH1 reproduces this transition to form amyloids. RepA-WH1 assembles double helical filaments by lateral association of a single-stranded precursor built by monomers. Double filaments then associate in mature fibers. The intracellular and cytotoxic RepA-WH1 aggregates might reproduce the hierarchical assembly of human amyloidogenic proteins. |
 External links External links |  Structure / Structure /  PubMed:25543255 PubMed:25543255 |
| Methods | EM (helical sym.) |
| Resolution | 29.0 Å |
| Structure data |  EMDB-6182: 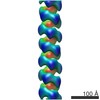 EMDB-6183: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




