+Search query
-Structure paper
| Title | Structural basis of prostaglandin efflux by MRP4. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 31, Issue 4, Page 621-632, Year 2024 |
| Publish date | Jan 12, 2024 |
 Authors Authors | Sergei Pourmal / Evan Green / Ruchika Bajaj / Ilan E Chemmama / Giselle M Knudsen / Meghna Gupta / Andrej Sali / Yifan Cheng / Charles S Craik / Deanna L Kroetz / Robert M Stroud /  |
| PubMed Abstract | Multidrug resistance protein 4 (MRP4) is a broadly expressed ATP-binding cassette transporter that is unique among the MRP subfamily for transporting prostanoids, a group of signaling molecules ...Multidrug resistance protein 4 (MRP4) is a broadly expressed ATP-binding cassette transporter that is unique among the MRP subfamily for transporting prostanoids, a group of signaling molecules derived from unsaturated fatty acids. To better understand the basis of the substrate selectivity of MRP4, we used cryogenic-electron microscopy to determine six structures of nanodisc-reconstituted MRP4 at various stages throughout its transport cycle. Substrate-bound structures of MRP4 in complex with PGE, PGE and the sulfonated-sterol DHEA-S reveal a common binding site that accommodates a diverse set of organic anions and suggest an allosteric mechanism for substrate-induced enhancement of MRP4 ATPase activity. Our structure of a catalytically compromised MRP4 mutant bound to ATP-Mg is outward-occluded, a conformation previously unobserved in the MRP subfamily and consistent with an alternating-access transport mechanism. Our study provides insights into the endogenous function of this versatile efflux transporter and establishes a basis for MRP4-targeted drug design. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:38216659 / PubMed:38216659 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.7 - 3.5 Å |
| Structure data | EMDB-40821, PDB-8swn: EMDB-40826, PDB-8sx7: EMDB-40827, PDB-8sx8: EMDB-40828, PDB-8sx9: EMDB-40829, PDB-8sxa: EMDB-40830, PDB-8sxb: |
| Chemicals |  ChemComp-MG:  ChemComp-PTY:  ChemComp-ATP: 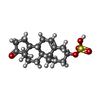 ChemComp-ZWY:  ChemComp-HOH: 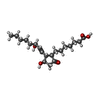 ChemComp-XPG: 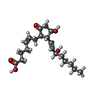 ChemComp-P2E: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / ABC transporter / multidrug resistance-associated protein / membrane protein |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers
















