+Search query
-Structure paper
| Title | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 2758, Year 2022 |
| Publish date | May 19, 2022 |
 Authors Authors | Injae Chung / John J Wright / Hannah R Bridges / Bozhidar S Ivanov / Olivier Biner / Caroline S Pereira / Guilherme M Arantes / Judy Hirst /    |
| PubMed Abstract | Mitochondrial complex I is a central metabolic enzyme that uses the reducing potential of NADH to reduce ubiquinone-10 (Q) and drive four protons across the inner mitochondrial membrane, powering ...Mitochondrial complex I is a central metabolic enzyme that uses the reducing potential of NADH to reduce ubiquinone-10 (Q) and drive four protons across the inner mitochondrial membrane, powering oxidative phosphorylation. Although many complex I structures are now available, the mechanisms of Q reduction and energy transduction remain controversial. Here, we reconstitute mammalian complex I into phospholipid nanodiscs with exogenous Q. Using cryo-EM, we reveal a Q molecule occupying the full length of the Q-binding site in the 'active' (ready-to-go) resting state together with a matching substrate-free structure, and apply molecular dynamics simulations to propose how the charge states of key residues influence the Q binding pose. By comparing ligand-bound and ligand-free forms of the 'deactive' resting state (that require reactivating to catalyse), we begin to define how substrate binding restructures the deactive Q-binding site, providing insights into its physiological and mechanistic relevance. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:35589726 / PubMed:35589726 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.3 - 3.02 Å |
| Structure data | EMDB-14132, PDB-7qsk: EMDB-14133, PDB-7qsl: EMDB-14134, PDB-7qsm: EMDB-14139, PDB-7qsn: EMDB-14140, PDB-7qso: |
| Chemicals |  ChemComp-3PE:  ChemComp-PC1:  ChemComp-SF4:  ChemComp-U10:  ChemComp-FES:  ChemComp-FMN:  ChemComp-K:  ChemComp-CDL:  ChemComp-GTP:  ChemComp-MG:  ChemComp-NDP:  ChemComp-ZN:  ChemComp-EHZ: 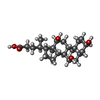 ChemComp-CHD:  ChemComp-MYR:  ChemComp-HOH:  ChemComp-GOL:  ChemComp-LMT: |
| Source |
|
 Keywords Keywords | OXIDOREDUCTASE / Mitochondrial complex I / Respiratory complex I / NADH:ubiquinone oxidoreductase / Ubiquinone / Nanodisc / ELECTRON TRANSPORT |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers