+Search query
-Structure paper
| Title | Structural and dynamic mechanisms of GABA receptor modulators with opposing activities. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 4582, Year 2022 |
| Publish date | Aug 6, 2022 |
 Authors Authors | Shaotong Zhu / Akshay Sridhar / Jinfeng Teng / Rebecca J Howard / Erik Lindahl / Ryan E Hibbs /   |
| PubMed Abstract | γ-Aminobutyric acid type A (GABA) receptors are pentameric ligand-gated ion channels abundant in the central nervous system and are prolific drug targets for treating anxiety, sleep disorders and ...γ-Aminobutyric acid type A (GABA) receptors are pentameric ligand-gated ion channels abundant in the central nervous system and are prolific drug targets for treating anxiety, sleep disorders and epilepsy. Diverse small molecules exert a spectrum of effects on γ-aminobutyric acid type A (GABA) receptors by acting at the classical benzodiazepine site. They can potentiate the response to GABA, attenuate channel activity, or counteract modulation by other ligands. Structural mechanisms underlying the actions of these drugs are not fully understood. Here we present two high-resolution structures of GABA receptors in complex with zolpidem, a positive allosteric modulator and heavily prescribed hypnotic, and DMCM, a negative allosteric modulator with convulsant and anxiogenic properties. These two drugs share the extracellular benzodiazepine site at the α/γ subunit interface and two transmembrane sites at β/α interfaces. Structural analyses reveal a basis for the subtype selectivity of zolpidem that underlies its clinical success. Molecular dynamics simulations provide insight into how DMCM switches from a negative to a positive modulator as a function of binding site occupancy. Together, these findings expand our understanding of how GABA receptor allosteric modulators acting through a common site can have diverging activities. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:35933426 / PubMed:35933426 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.9 Å |
| Structure data | EMDB-27332, PDB-8dd2: EMDB-27333, PDB-8dd3: |
| Chemicals |  ChemComp-NAG: 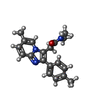 ChemComp-R5R:  ChemComp-ABU: 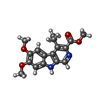 ChemComp-R63: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN/IMMUNE SYSTEM / GABAA receptor / Zolpidem / MEMBRANE PROTEIN-IMMUNE SYSTEM complex / Human GABAA receptor / DMCM |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








 homo sapiens (human)
homo sapiens (human)