+Search query
-Structure paper
| Title | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. |
|---|---|
| Journal, issue, pages | Cell, Vol. 184, Issue 2, Page 370-383.e13, Year 2021 |
| Publish date | Jan 21, 2021 |
 Authors Authors | Nan Wang / Xin Jiang / Shuo Zhang / Angqi Zhu / Yafei Yuan / Hanwen Xu / Jianlin Lei / Chuangye Yan /   |
| PubMed Abstract | Proton-coupled monocarboxylate transporters MCT1-4 catalyze the transmembrane movement of metabolically essential monocarboxylates and have been targeted for cancer treatment because of their ...Proton-coupled monocarboxylate transporters MCT1-4 catalyze the transmembrane movement of metabolically essential monocarboxylates and have been targeted for cancer treatment because of their enhanced expression in various tumors. Here, we report five cryo-EM structures, at resolutions of 3.0-3.3 Å, of human MCT1 bound to lactate or inhibitors in the presence of Basigin-2, a single transmembrane segment (TM)-containing chaperon. MCT1 exhibits similar outward-open conformations when complexed with lactate or the inhibitors BAY-8002 and AZD3965. In the presence of the inhibitor 7ACC2 or with the neutralization of the proton-coupling residue Asp309 by Asn, similar inward-open structures were captured. Complemented by structural-guided biochemical analyses, our studies reveal the substrate binding and transport mechanism of MCTs, elucidate the mode of action of three anti-cancer drug candidates, and identify the determinants for subtype-specific sensitivities to AZD3965 by MCT1 and MCT4. These findings lay out an important framework for structure-guided drug discovery targeting MCTs. |
 External links External links |  Cell / Cell /  PubMed:33333023 PubMed:33333023 |
| Methods | EM (single particle) |
| Resolution | 2.95 - 3.3 Å |
| Structure data | EMDB-30019, PDB-6lyy: EMDB-30020, PDB-6lz0: EMDB-30389: Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate 7ACC2 in the inward-open conformation. EMDB-30391, PDB-7ckr: EMDB-30623, PDB-7da5: |
| Chemicals | 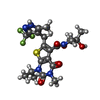 ChemComp-EY0: 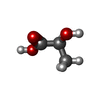 ChemComp-2OP:  ChemComp-G5L:  ChemComp-G5O: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / Proton-coupled monocarboxylate transporter / MCT1 / Basigin / anti-cancer / AZD3965 / BAY-8002 / 7ACC2 / single particle cryo-EM / latate transporter / lactate transporter / Basigin-2 / anti-cancer drug candidate |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













 homo sapiens (human)
homo sapiens (human)