+検索条件
-Structure paper
| タイトル | A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody. |
|---|---|
| ジャーナル・号・ページ | bioRxiv, Year 2020 |
| 掲載日 | 2020年9月21日 |
 著者 著者 | Nicholas C Wu / Meng Yuan / Sandhya Bangaru / Deli Huang / Xueyong Zhu / Chang-Chun D Lee / Hannah L Turner / Linghang Peng / Linlin Yang / David Nemazee / Andrew B Ward / Ian A Wilson /  |
| PubMed 要旨 | Epitopes that are conserved among SARS-like coronaviruses are attractive targets for design of cross-reactive vaccines and therapeutics. CR3022 is a SARS-CoV neutralizing antibody to a highly ...Epitopes that are conserved among SARS-like coronaviruses are attractive targets for design of cross-reactive vaccines and therapeutics. CR3022 is a SARS-CoV neutralizing antibody to a highly conserved epitope on the receptor binding domain (RBD) on the spike protein that can cross-react with SARS-CoV-2, but with lower affinity. Using x-ray crystallography, mutagenesis, and binding experiments, we illustrate that of four amino acid differences in the CR3022 epitope between SARS-CoV-2 and SARS-CoV, a single mutation P384A fully determines the affinity difference. CR3022 does not neutralize SARS-CoV-2, but the increased affinity to SARS-CoV-2 P384A mutant now enables neutralization with a similar potency to SARS-CoV. We further investigated CR3022 interaction with the SARS-CoV spike protein by negative-stain EM and cryo-EM. Three CR3022 Fabs bind per trimer with the RBD observed in different up-conformations due to considerable flexibility of the RBD. In one of these conformations, quaternary interactions are made by CR3022 to the N-terminal domain (NTD) of an adjacent subunit. Overall, this study provides insights into antigenic variation and potential for cross-neutralizing epitopes on SARS-like viruses. |
 リンク リンク |  bioRxiv / bioRxiv /  PubMed:32995788 / PubMed:32995788 /  PubMed Central PubMed Central |
| 手法 | EM (単粒子) |
| 解像度 | 6.15 - 24.0 Å |
| 構造データ |  EMDB-22858: 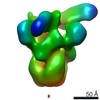 EMDB-22859: 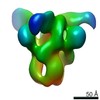 EMDB-22860:  EMDB-22861:  EMDB-22862:  EMDB-22863:  EMDB-22864: |
| 由来 |
|
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア 万見文献について
万見文献について




