-Search query
Query structure data:
- EMDB map
- 4 or 5 digit ID
- or string starts with "e"
- e.g. 1003, EMD-1003, e1003
- PDB deposited unit (asymetric unit, AU)
- 4 letter ID code
- or ID code + hyphen + "d"
- e.g. 100d, 100d-d
- PDB assembly (biological unit, BU)
- ID + hyphen + assembly-ID
- Or click the image of AU/BU, which will be shown below when you input an PDB-ID
- e.g. 1oel-1
- SASBDB model
- SASBDB-ID
- or SASBDB-model-ID
- e.g. SASDAA5, 100, sas-100
 Comparison of 3 databanks
Comparison of 3 databanksSamples:

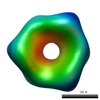
This is an EM map data of the RNA polymerase II at 25 A resolution. You will find atomic models and EM maps of RNA polymerase structures from the EMDB & PDB._Perform search.

The DNA clamp is a ring shaped oligomer essential for DNA metabolism. All the DNA clamps have similar shapes, while bacterial clamps are dimer ring, and archaeal/eukaryal ones are trimer ring. By search using this trimer clamp, you will find dimer clamp structures as well._Perform search.

EF-Tu/tRNA complex (RNA + protein) and EF-G (monomeric protein) have similar shapes despite their different compositions ("molecular mimicry"). You will find both EFs by this search._Perform search.

This is a dummy-atom model of lumazine synthase obtained by SAS. You will find many crystal structures and EM maps of same molecules._Perform search.

Many 70S ribosome structure data are stored in EMDB and PDB. Omokage search is useful to find out such the huge molecule structures._Perform search.

For huger molecule, such as this rice dwarf virus, Omokage search can be used._Perform search.

Not only for the huge structures but also for small molecules, such as this DNA fragment, this tool might be useful. By this search, some proteins having similar shape to this DNA fragment are found (for good or for bad)._Perform search.

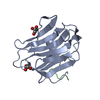
This is another example for small molecules. Which proteins have simialar shapes to this beta-propeller structure?_Perform search.
-Query data not found
- About Omokage search
About Omokage search
-News
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
-Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jul 12, 2017. Major update of PDB
+Jun 16, 2017. Omokage search with filter
+Apr 13, 2016. Omokage search got faster
-Omokage search
Search structure by SHAPE
- "Omokage search" is a shape similarity search service for 3D structures of macromolecules. By comparing global shapes, and ignoring details, similar-shaped structures are searched.
- The search is performed ageinst >200,000 structure data, which consists of EMDB map data, PDB coordinates (deposited units (asymmetric units, usually), PDB biological units, and SASBDB mdoels).
- For the search query, you can use either a data in the PDB/EMDB/SASBDB or your original model.
- Supported formats are PDB (atomic model, SAXS bead model, etc.) and CCP4/MRC map (3D density map).
- Shape comparison is performed by iDR profile method, which uses 1D-distance profiles of super-simplified models generated by vector quantizaion in Situs package.
Related info.: Comparison of 3 databanks /
Comparison of 3 databanks /  Sep 22, 2014. New service: Omokage search /
Sep 22, 2014. New service: Omokage search /  gmfit /
gmfit /  Re-ranking by gmfit
Re-ranking by gmfit
 Movie
Movie Controller
Controller Structure viewers
Structure viewers