[English] 日本語
 Yorodumi
Yorodumi- EMDB-9030: HIV-1 Envelope Glycoprotein Clone B41_delCT in complex with the b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9030 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 Envelope Glycoprotein Clone B41_delCT in complex with the broadly neutralizing antibody Fab PGT151 | |||||||||
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.7 Å | |||||||||
 Authors Authors | Berndsen ZT / Ward AB | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Authors: Liwei Cao / Matthias Pauthner / Raiees Andrabi / Kimmo Rantalainen / Zachary Berndsen / Jolene K Diedrich / Sergey Menis / Devin Sok / Raiza Bastidas / Sung-Kyu Robin Park / Claire M ...Authors: Liwei Cao / Matthias Pauthner / Raiees Andrabi / Kimmo Rantalainen / Zachary Berndsen / Jolene K Diedrich / Sergey Menis / Devin Sok / Raiza Bastidas / Sung-Kyu Robin Park / Claire M Delahunty / Lin He / Javier Guenaga / Richard T Wyatt / William R Schief / Andrew B Ward / John R Yates / Dennis R Burton / James C Paulson /  Abstract: As the sole target of broadly neutralizing antibodies (bnAbs) to HIV, the envelope glycoprotein (Env) trimer is the focus of vaccination strategies designed to elicit protective bnAbs in humans. ...As the sole target of broadly neutralizing antibodies (bnAbs) to HIV, the envelope glycoprotein (Env) trimer is the focus of vaccination strategies designed to elicit protective bnAbs in humans. Because HIV Env is densely glycosylated with 75-90 N-glycans per trimer, most bnAbs use or accommodate them in their binding epitope, making the glycosylation of recombinant Env a key aspect of HIV vaccine design. Upon analysis of three HIV strains, we here find that site-specific glycosylation of Env from infectious virus closely matches Envs from corresponding recombinant membrane-bound trimers. However, viral Envs differ significantly from recombinant soluble, cleaved (SOSIP) Env trimers, strongly impacting antigenicity. These results provide a benchmark for virus Env glycosylation needed for the design of soluble Env trimers as part of an overall HIV vaccine strategy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9030.map.gz emd_9030.map.gz | 116.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9030-v30.xml emd-9030-v30.xml emd-9030.xml emd-9030.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
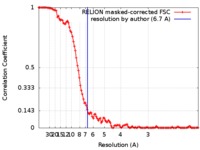
| FSC (resolution estimation) |  emd_9030_fsc.xml emd_9030_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_9030.png emd_9030.png | 59.2 KB | ||
| Others |  emd_9030_half_map_1.map.gz emd_9030_half_map_1.map.gz emd_9030_half_map_2.map.gz emd_9030_half_map_2.map.gz | 98.4 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9030 http://ftp.pdbj.org/pub/emdb/structures/EMD-9030 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9030 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9030 | HTTPS FTP |
-Validation report
| Summary document |  emd_9030_validation.pdf.gz emd_9030_validation.pdf.gz | 79 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9030_full_validation.pdf.gz emd_9030_full_validation.pdf.gz | 78.1 KB | Display | |
| Data in XML |  emd_9030_validation.xml.gz emd_9030_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9030 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9030 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9030 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9030 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9030.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9030.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
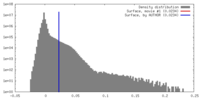
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1
| File | emd_9030_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
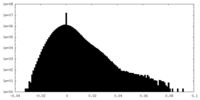
| Projections & Slices |
| ||||||||||||
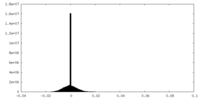
| Density Histograms |
-Half map: Half Map 2
| File | emd_9030_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
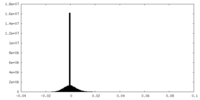
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Envelope Glycoprotein Clone B41dCT in complex with the broa...
| Entire | Name: HIV-1 Envelope Glycoprotein Clone B41dCT in complex with the broadly neutralizing antibody Fab PGT151 |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Envelope Glycoprotein Clone B41dCT in complex with the broa...
| Supramolecule | Name: HIV-1 Envelope Glycoprotein Clone B41dCT in complex with the broadly neutralizing antibody Fab PGT151 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: HIV-1 Envelope Glycoprotein Clone B41_delCT
| Macromolecule | Name: HIV-1 Envelope Glycoprotein Clone B41_delCT / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAAKKW VTVYYGVPVW KEATTT LFC ASDAKAYDTE VHNVWATHAC VPTDPNPQEI VLGNVTENFN MWKNNMVEQM HEDIISL WD QSLKPCVKLT PLCVTLNCNN VNTNNTNNST NATISDWEKM ETGEMKNCSF ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAAKKW VTVYYGVPVW KEATTT LFC ASDAKAYDTE VHNVWATHAC VPTDPNPQEI VLGNVTENFN MWKNNMVEQM HEDIISL WD QSLKPCVKLT PLCVTLNCNN VNTNNTNNST NATISDWEKM ETGEMKNCSF NVTTSIRD K IKKEYALFYK LDVVPLENKN NINNTNITNY RLINCNTSVI TQACPKVSFE PIPIHYCAP AGFAILKCNS KTFNGSGPCT NVSTVQCTHG IRPVVSTQLL LNGSLAEEEI VIRSENITDN AKTIIVQLN EAVEINCTRP NNNTRKSIHI GPGRAFYATG DIIGNIRQAH CNISKARWNE T LGQIVAKL EEQFPNKTII FNHSSGGDPE IVTHSFNCGG EFFYCNTTPL FNSTWNNTRT DD YPTGGEQ NITLQCRIKQ IINMWQGVGK AMYAPPIRGQ IRCSSNITGL LLTRDGGRDQ NGT ETFRPG GGNMRDNWRS ELYKYKVVKI EPLGIAPTAA KRRVVQREKR AVGLGAFILG FLGA AGSTM GAASMALTVQ ARLLLSGIVQ QQNNLLRAIE AQQHMLQLTV WGIKQLQARV LAVER YLRD QQLLGIWGCS GKIICTTNVP WNDSWSNKTI NEIWDNMTWM QWEKEIDNYT QHIYTL LEV SQIQQEKNEQ ELLELDKWDS LWNWFSISNW LWYIKIFIMI VGGLIGLRIV FTVLSII SR VRQGGGSGGG WSHPQFEK |
-Macromolecule #2: Immunoglobulin G PGT151 Fab, Heavy chain
| Macromolecule | Name: Immunoglobulin G PGT151 Fab, Heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RVQLVESGGG VVQPGKSVRL SCVVSDFPFS KYPMYWVRQA PGKGLEWVAA ISGDAWHVVY SNSVQGRFLV SRDNVKNTLY LEMNSLKIE DTAVYRCARM FQESGPPRLD RWSGRNYYYY SGMDVWGQGT TVTVSSASTK GPSVFPLAPS SKSTSGGTAA L GCLVKDYF ...String: RVQLVESGGG VVQPGKSVRL SCVVSDFPFS KYPMYWVRQA PGKGLEWVAA ISGDAWHVVY SNSVQGRFLV SRDNVKNTLY LEMNSLKIE DTAVYRCARM FQESGPPRLD RWSGRNYYYY SGMDVWGQGT TVTVSSASTK GPSVFPLAPS SKSTSGGTAA L GCLVKDYF PEPVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKRVEPKSC DK |
-Macromolecule #3: Immunoglobulin G PGT151, Light chain
| Macromolecule | Name: Immunoglobulin G PGT151, Light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Sequence | String: DIVMTQTPLS LSVTPGQPAS ISCKSSESLR QSNGKTSLYW YRQKPGQSPQ LLVFEVSNRF SGVSDRFVGS GSGTDFTLRI SRVEAEDVG FYYCMQSKDF PLTFGGGTKV DLKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: DIVMTQTPLS LSVTPGQPAS ISCKSSESLR QSNGKTSLYW YRQKPGQSPQ LLVFEVSNRF SGVSDRFVGS GSGTDFTLRI SRVEAEDVG FYYCMQSKDF PLTFGGGTKV DLKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.13 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2238 / Average exposure time: 8.5 sec. / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)