[English] 日本語
 Yorodumi
Yorodumi- EMDB-8303: Structure of rabbit RyR2 in complex with FKBP12.6 in a closed sta... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8303 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of rabbit RyR2 in complex with FKBP12.6 in a closed state (conformation C1) | |||||||||
 Map data Map data | Ryanodine Receptor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Calcium Release Channel / Ryanodine Receptor / TRANSPORT PROTEIN-ISOMERASE complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of sequestering of calcium ion / cyclic nucleotide binding / negative regulation of calcium-mediated signaling / negative regulation of insulin secretion involved in cellular response to glucose stimulus / negative regulation of release of sequestered calcium ion into cytosol / neuronal action potential propagation / insulin secretion involved in cellular response to glucose stimulus / response to redox state / 'de novo' protein folding / negative regulation of heart rate ...positive regulation of sequestering of calcium ion / cyclic nucleotide binding / negative regulation of calcium-mediated signaling / negative regulation of insulin secretion involved in cellular response to glucose stimulus / negative regulation of release of sequestered calcium ion into cytosol / neuronal action potential propagation / insulin secretion involved in cellular response to glucose stimulus / response to redox state / 'de novo' protein folding / negative regulation of heart rate / FK506 binding / positive regulation of axon regeneration / smooth muscle contraction / response to vitamin E / calcium channel inhibitor activity / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / T cell proliferation / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / Ion homeostasis / release of sequestered calcium ion into cytosol / sarcoplasmic reticulum membrane / calcium channel regulator activity / calcium channel complex / protein maturation / peptidyl-prolyl cis-trans isomerase activity / RNA polymerase II CTD heptapeptide repeat P3 isomerase activity / RNA polymerase II CTD heptapeptide repeat P6 isomerase activity / peptidylprolyl isomerase / calcium-mediated signaling / response to hydrogen peroxide / Stimuli-sensing channels / Z disc / positive regulation of cytosolic calcium ion concentration / protein refolding / transmembrane transporter binding / signaling receptor binding / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.0 Å | |||||||||
 Authors Authors | Lobo JJ / Dhindwal S / Samso M | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Signal / Year: 2017 Journal: Sci Signal / Year: 2017Title: A cryo-EM-based model of phosphorylation- and FKBP12.6-mediated allosterism of the cardiac ryanodine receptor. Authors: Sonali Dhindwal / Joshua Lobo / Vanessa Cabra / Demetrio J Santiago / Ashok R Nayak / Kelly Dryden / Montserrat Samsó /  Abstract: Type 2 ryanodine receptors (RyR2s) are calcium channels that play a vital role in triggering cardiac muscle contraction by releasing calcium from the sarcoplasmic reticulum into the cytoplasm. ...Type 2 ryanodine receptors (RyR2s) are calcium channels that play a vital role in triggering cardiac muscle contraction by releasing calcium from the sarcoplasmic reticulum into the cytoplasm. Several cardiomyopathies are associated with the abnormal functioning of RyR2. We determined the three-dimensional structure of rabbit RyR2 in complex with the regulatory protein FKBP12.6 in the closed state at 11.8 Å resolution using cryo-electron microscopy and built an atomic model of RyR2. The heterogeneity in the data set revealed two RyR2 conformations that we proposed to be related to the extent of phosphorylation of the P2 domain. Because the more flexible conformation may correspond to RyR2 with a phosphorylated P2 domain, we suggest that phosphorylation may set RyR2 in a conformation that needs less energy to transition to the open state. Comparison of RyR2 from cardiac muscle and RyR1 from skeletal muscle showed substantial structural differences between the two, especially in the helical domain 2 (HD2) structure forming the Clamp domain, which participates in quaternary interactions with the dihydropyridine receptor and neighboring RyRs in RyR1 but not in RyR2. Rigidity of the HD2 domain of RyR2 was enhanced by binding of FKBP12.6, a ligand that stabilizes RyR2 in the closed state. These results help to decipher the molecular basis of the different mechanisms of activation and oligomerization of the RyR isoforms and could be extended to RyR complexes in other tissues. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8303.map.gz emd_8303.map.gz | 136.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8303-v30.xml emd-8303-v30.xml emd-8303.xml emd-8303.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8303_1.png emd_8303_1.png emd_8303_2.png emd_8303_2.png | 128.8 KB 97.9 KB | ||
| Filedesc metadata |  emd-8303.cif.gz emd-8303.cif.gz | 9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8303 http://ftp.pdbj.org/pub/emdb/structures/EMD-8303 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8303 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8303 | HTTPS FTP |
-Validation report
| Summary document |  emd_8303_validation.pdf.gz emd_8303_validation.pdf.gz | 399.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8303_full_validation.pdf.gz emd_8303_full_validation.pdf.gz | 398.9 KB | Display | |
| Data in XML |  emd_8303_validation.xml.gz emd_8303_validation.xml.gz | 7.3 KB | Display | |
| Data in CIF |  emd_8303_validation.cif.gz emd_8303_validation.cif.gz | 8.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8303 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8303 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8303 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8303 | HTTPS FTP |
-Related structure data
| Related structure data |  5l1dMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8303.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8303.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ryanodine Receptor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
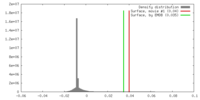
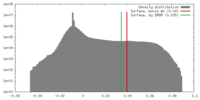
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ryanodine Receptor - FKBP12.6
| Entire | Name: Ryanodine Receptor - FKBP12.6 |
|---|---|
| Components |
|
-Supramolecule #1: Ryanodine Receptor - FKBP12.6
| Supramolecule | Name: Ryanodine Receptor - FKBP12.6 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.26 MDa |
-Macromolecule #1: Ryanodine Receptor
| Macromolecule | Name: Ryanodine Receptor / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 487.120906 KDa |
| Sequence | String: MADGGEGEDE IQFLRTDDEV VLQCTATIHK EQQKLCLAAE GFGNRLCFLE STSNSKNVPP DLSICTFVLE QSLLVRALQE MLANTVEKS EGQVDVEKWK FMMKTAQGGG HRTLLYGHAI LLRHSYSGMY LCCLSTSRSS TDKLAFDVGL QEDTTGEACW W TIHPASKQ ...String: MADGGEGEDE IQFLRTDDEV VLQCTATIHK EQQKLCLAAE GFGNRLCFLE STSNSKNVPP DLSICTFVLE QSLLVRALQE MLANTVEKS EGQVDVEKWK FMMKTAQGGG HRTLLYGHAI LLRHSYSGMY LCCLSTSRSS TDKLAFDVGL QEDTTGEACW W TIHPASKQ RSEGEKVRVG DDLILVSVSS ERYLHLSYGN GSLHVDAAFQ QTLWSVAPIS SGSEAAQGYL IGGDVLRLLH GH MDECLTV PSGEHGEEQR RTVHYEGGAV SVHARSLWRL ETLRVAWSGS HIRWGQPFRL RHVTTGKYLS LMEDKNLLLM DKE KADVKS TAFTFRSSKE KLDGGVRKEV DGMGTSEIKY GDSICYIQHV DTGLWLTYQS VDVKSVRMGS IQRKAIMHHE GHMD DGLNL SRSQHEESRT ARVIRSTVFL FNRFIRGLDA LSKKAKASSV DLPIESVSLS LQDLIGYFHP PDEHLEHEDK QNRLR ALKN RQNLFQEEGM INLVLECIDR LHVYSSAAHF ADVAGREAGE SWKSILNSLY ELLAALIRGN RKNCAQFSGS LDWLIS RLE RLEASSGILE VLHCVLVESP EALNIIKEGH IKSIISLLDK HGRNHKVLDV LCSLCVCHGV AVRSNQHLIC DNLLPGR DL LLQTRLVNHV SSMRPNIFLG VSEGSAQYKK WYYELMVDHT EPFVTAEATH LRVGWASTEG YSPYPGGGEE WGGNGVGD D LFSYGFDGLH LWSGCIARTV SSPNQHLLRT DDVISCCLDL SAPSISFRIN GQPVQGMFEN FNIDGLFFPV VSFSAGIKV RFLLGGRHGE FKFLPPPGYA PCYEAVLPKE KLKVEHSREY KQERTYTRDL LGPTVSLTQA AFTPIPVDTS QIVLPPHLER IREKLAENI HELWVMNKIE LGWQYGPVRD DNKRQHPCLV EFSKLPEQER NYNLQMSLET LKTLLALGCH VGISDEHAEE K VKKMKLPK NYQLTSGYKP APMDLSFIKL TPSQEAMVDK LAENAHNVWA RDRIRQGWTY GIQQDVKNRR NPRLVPYTLL DD RTKKSNK DSLREAVRTL LGYGYNLEAP DQDHAARAEV CSGTGERFRI FRAEKTYAVK AGRWYFEFEA VTSGDMRVGW SRP GCQPDQ ELGSDERAFA FDGFKAQRWH QGNEHYGRSW QAGDVVGCMV DMNEHTMMFT LNGEILLDDS GSELAFKDFD VGDG FIPVC SLGVAQVGRM NFGKDVSTLK YFTICGLQEG YEPFAVNTNR DITMWLSKRL PQFLQVPSNH EHIEVTRIDG TIDSS PCLK VTQKSFGSQN SNTDIMFYRL SMPIECAEVF SKTVPGGLPG AGLFGPKNDL EDYDADSDFE VLMKTAHGHL VPDRVD KDK ETTKAEFNNH KDYAQEKPSR LKQRFLLRRT KPDYSTSHSA RLTEDVLADD RDDYDFLMQT STYYYSVRIF PGQEPAN VW VGWITSDFHQ YDTGFDLDRV RTVTVTLGDE KGKVHESIKR SN(UNK)(UNK)(UNK)(UNK)(UNK)NNN GLEIGCVV D AASGLLTFIA NGKELSTYYQ VEPSTKLFPA VFAQATSPNV FQFELGRIKN VMPLSAGLFK SEHKNPVPQC PPRLHVQFL SHVLWSRMPN QFLKVDVSRI SERQGWLVQC LDPLQFMSLH IPEENRSVDI LELTEQEELL KFHYHTLRLY SAVCALGNHR VAHALCSHV DEPQLLYAIE NKYMPGLLRT GYYDLLIDIH LSSYATARLM MNNEFIVPMT EETKSITLFP DENKKHGLPG I GLSTSLRP RMQFSSPSFV SINNECYQYS PEFPLDILKA KTIQMLTEAV KEGSLHGRDP VGGTTEFLFV PLIKLFYTLL IM GIFHNED LRHILQLIEP SVFKDAATPE EEGDTLEEEP SVEDTKLEGA GEEEAKVGKR PKEGLLQMKL PEPVKLQMCL LLQ YLCDCQ VRHRIEAIVA FSDDFVAKLQ DNQRFR(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)LTIRGRLL SLVEKVTYLK KKQTEKPVES DSRKSSTLQQ LISETMVRWA QE SVIEDPE LVRAMFVLLH RQYDGIGGLV RALPKTYTIN GVSVEDTINL LASLGQIRSL LSVRMGKEEE KLMIRGLGDI MNN KVFYQH PNLMRALGMH ETVMEVMVNV LGGGESKEIT FPKMVANCCR FLCYFCRISR QNQKAMFDHL SYLLENSSVG LASP AMRGS TPLDVAAASV MDNNELALAL REPDLEKVVV RYLAGCGLQS CQMLVSKGYP DIGWNPVEGE RYLDFLRFAV FCNGE SVEE NANVVVRLLI RRPECFGPAL RGEGGNGLLA AMEEAIKIAE DPSRDGPSPT SGSSKTLDTE EEEDDTIHMG NAIMTF YAA LIDLLGRCAP EMHLIHAGKG EAIRIRSILR SL(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)FCP DHKAAMVLFL DRVYGIEVQD FLLHLLEVGF LPDLRAAASL DTAALSATDM AL ALNRYLC TAVLPLL(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)FN PQP VDTSNI IIPEKLEYFI NKYAEHSHDK WSMDKLANGW IYGEIYSDSS KIQPLMKPYK LLSEKEKEIY RWPIKESLKT MLAW GWRIE RTREGDSMAL YNRTRRISQT SQVSVDAAHG YSPRAIDMSN VTLSRDLHAM AEMMAENYHN IWAKKKKLEL ESKGG GNHP LLVPYDTLTA KEKAKDREKA QDILKFLQIN GYAVSRG(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)P RHRAVNLFLQ GYEKSWIETE EHYFEDKLIE DLAKPGAEPP EEDEVTKRVD PLHQLILLFS RTALTEKCK LEEDFLYMAY ADIMAKSCHD EEDDDGEEEV KSFEEKEMEK QKLLYQQARL HDRGAAEMVL QTISASKGET GPMVAATLKL GIAILNGGN STVQQKMLDY LKEKKDVGFF QSLAGLMQSC SVLDLNAFER QNKAEGLGMV TEEGSGEKVL QDDEFTCDLF R FLQLLCEG HNSDFQNYLR TQTGNNTTVN IIISTVDYLL RVQESISDFY WYYSGKDVID EQGQRNFSKA IQVAKQVFNT LT EYIQGPC TGNQQSLAHS RLWDAVVGFL HVFAHMQMKL SQDSSQIELL KELMDLQKDM VVMLLSMLEG NVVNGTIGKQ MVD MLVESS NNVEMILKFF DMFLKLKDLT SSDTFKEYDP DGKGIISKRD FHKAMESHKH YTQSETEFLL SCAETDENET LDYE EFVKR FHEPAKDIGF NVAVLLTNLS EHMPNDTRLQ TFLELAESVL NYFQPFLGRI EIMGSAKRIE RVYFEISESS RTQWE KPQV KESKRQFIFD VVNEGGEKEK MELFVNFCED TIFEMQLAAQ ISESDLNERS ANKEESEKER PEEQGPKMGF FSVLTV RSA LFALRYNILT LMRMLSLKSL KKQMKKMKKM TVKDMVTAFF SSYWSIFMTL LHFVASVFRG FFRIVCSLLL GGSLVEG AK KIKVAELLAN MPDPTQDEVR GDGEEGERKP METTLPSEDL TDLKELTEES DLLSDIFGLD LKREGGQYKL IPHNPNAG L SDLMSNPVLI PEEQEKFQEQ KTKEEEKEEK EETKSEPEKA EGEDGEKEEK VKEDKGKQKL RQLHTHRYGE PEVPESAFW KKIIAYQQKL LNYLARNFYN MRMLALFVAF AINFILLFYK VSTSSVVEGK ELPSRSTSEN AKVTTSLDSS SHRIIAVHYV LEESSGYME PTLRILAILH TVISFFCIIG YYCLKVPLVI FKREKEVARK LEFDGLYITE QPSEDDIKGQ WDRLVINTQS F PNNYWDKF VKRKVMDKYG EFYGRDRISE LLGMDKAALD FSDAREKKKP KKDSSLSAVL NSIDVKYQMW KLGVVFTDNS FL YLAWYMT MSILGHYNNF FFAAHLLDIA MGFKTLRTIL SSVTHNGKQL VLTVGLLAVV VYLYTVVAFN FFRKFYNKSE DGD TPDMKC DDMLTCYMFH MYVGVRAGGG IGDEIEDPAG DEYEIYRIIF DITFFFFVIV ILLAIIQGLI IDAFGELRDQ QEQV KEDME TKCFICGIGN DYFDTVPHGF ETHTLQEHNL ANYLFFLMYL INKDETEHTG QESYVWKMYQ ERCWEFFPAG DCFRK QYED QLN |
-Macromolecule #2: Peptidyl-prolyl cis-trans isomerase FKBP1B
| Macromolecule | Name: Peptidyl-prolyl cis-trans isomerase FKBP1B / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: peptidylprolyl isomerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.645984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNHKVHHHHH HMDEKTTGWR GGHVVEGLAG ELEQLRARLE HHPQGQREPE LGVEIETISP GDGRTFPKKG QTCVVHYTGM LQNGKKFDS SRDRNKPFKF RIGKQEVIKG FEEGAAQMSL GQRAKLTCTP DVAYGATGHP GVIPPNATLI FDVELLNLE UniProtKB: Peptidyl-prolyl cis-trans isomerase FKBP1B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 1x Protease Inhibitor cocktail | ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 20 | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295.15 K / Instrument: FEI VITROBOT MARK IV Details: Blot for 2 seconds before plunging into liquid ethane (FEI VITROBOT MARK IV).. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: OTHER / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-7 / Number grids imaged: 1 / Number real images: 858 / Average exposure time: 1.2 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 62000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5l1d: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)