[English] 日本語
 Yorodumi
Yorodumi- EMDB-8227: Tomographic subvolume average of membrane-bound Ebola (EBOV-Makon... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8227 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tomographic subvolume average of membrane-bound Ebola (EBOV-Makona) glycoprotein bound to c2G4 antibody | |||||||||
 Map data Map data | Membrane-bound Ebola (EBOV-Makona) glycoprotein bound to c2G4 antibody | |||||||||
 Sample Sample | virus-like particle / antibody complex != Ebola virus sp. virus-like particle / antibody complex
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Ebola virus sp. Ebola virus sp. | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 25.0 Å | |||||||||
 Authors Authors | Tran EEH / Subramaniam S | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2016 Journal: J Virol / Year: 2016Title: Mapping of Ebolavirus Neutralization by Monoclonal Antibodies in the ZMapp Cocktail Using Cryo-Electron Tomography and Studies of Cellular Entry. Authors: Erin E H Tran / Elizabeth A Nelson / Pranay Bonagiri / James A Simmons / Charles J Shoemaker / Connie S Schmaljohn / Gary P Kobinger / Larry Zeitlin / Sriram Subramaniam / Judith M White /   Abstract: ZMapp, a cocktail of three monoclonal antibodies (MAbs; c2G4, c4G7, and c13C6) against the ebolavirus (EBOV) glycoprotein (GP), shows promise for combatting outbreaks of EBOV, as occurred in West ...ZMapp, a cocktail of three monoclonal antibodies (MAbs; c2G4, c4G7, and c13C6) against the ebolavirus (EBOV) glycoprotein (GP), shows promise for combatting outbreaks of EBOV, as occurred in West Africa in 2014. Prior studies showed that Fabs from these MAbs bind a soluble EBOV GP ectodomain and that MAbs c2G4 and c4G7, but not c13C6, neutralize infections in cell cultures. Using cryo-electron tomography, we extended these findings by characterizing the structures of c2G4, c4G7, and c13C6 IgGs bound to native, full-length GP from the West African 2014 isolate embedded in filamentous viruslike particles (VLPs). As with the isolated ectodomain, c13C6 bound to the glycan cap, whereas c2G4 and c4G7 bound to the base region of membrane-bound GP. The tomographic data suggest that all three MAbs bind with high occupancy and that the base-binding antibodies can potentially bridge neighboring GP spikes. Functional studies indicated that c2G4 and c4G7, but not c13C6, competitively inhibit entry of VLPs bearing EBOV GP into the host cell cytoplasm, without blocking trafficking of VLPs to NPC1(+) endolysosomes, where EBOV fuses. Moreover, c2G4 and c4G7 bind to and can block entry mediated by the primed (19-kDa) form of GP without impeding binding of the C-loop of NPC1, the endolysosomal receptor for EBOV. The most likely mode of action of c2G4 and c4G7 is therefore by inhibiting conformational changes in primed, NPC1-bound GP that initiate fusion between the viral and target membranes, similar to the action of certain broadly neutralizing antibodies against influenza hemagglutinin and HIV Env. IMPORTANCE: The recent West African outbreak of ebolavirus caused the deaths of more than 11,000 individuals. Hence, there is an urgent need to be prepared with vaccines and therapeutics for similar ...IMPORTANCE: The recent West African outbreak of ebolavirus caused the deaths of more than 11,000 individuals. Hence, there is an urgent need to be prepared with vaccines and therapeutics for similar future disasters. ZMapp, a cocktail of three MAbs directed against the ebolavirus glycoprotein, is a promising anti-ebolavirus therapeutic. Using cryo-electron tomography, we provide structural information on how each of the MAbs in this cocktail binds to the ebolavirus glycoprotein as it is displayed-embedded in the membrane and present at high density-on filamentous viruslike particles that recapitulate the surface structure and entry functions of ebolavirus. Moreover, after confirming that two of the MAbs bind to the same region in the base of the glycoprotein, we show that they competitively block the entry function of the glycoprotein and that they can do so after the glycoprotein is proteolytically primed and bound to its intracellular receptor, Niemann-Pick C1. These findings should inform future developments of ebolavirus therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8227.map.gz emd_8227.map.gz | 637 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8227-v30.xml emd-8227-v30.xml emd-8227.xml emd-8227.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8227.png emd_8227.png | 42.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8227 http://ftp.pdbj.org/pub/emdb/structures/EMD-8227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8227 | HTTPS FTP |
-Validation report
| Summary document |  emd_8227_validation.pdf.gz emd_8227_validation.pdf.gz | 78.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8227_full_validation.pdf.gz emd_8227_full_validation.pdf.gz | 77.6 KB | Display | |
| Data in XML |  emd_8227_validation.xml.gz emd_8227_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8227 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8227 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8227 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8227 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8227.map.gz / Format: CCP4 / Size: 1.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8227.map.gz / Format: CCP4 / Size: 1.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Membrane-bound Ebola (EBOV-Makona) glycoprotein bound to c2G4 antibody | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
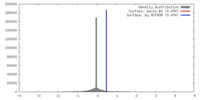
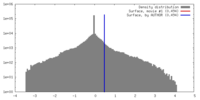
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : virus-like particle / antibody complex
| Entire | Name: virus-like particle / antibody complex |
|---|---|
| Components |
|
-Supramolecule #2: Ebola virus sp.
| Supramolecule | Name: Ebola virus sp. / type: virus / ID: 2 / Parent: 1 Details: 293T/17 cells were transfected with cDNAs encoding EBOV GP, VP40, mCherry-tagged VP40, and beta-lactamase-tagged VP40. After transfection, VLPs were purified from the cell medium by ...Details: 293T/17 cells were transfected with cDNAs encoding EBOV GP, VP40, mCherry-tagged VP40, and beta-lactamase-tagged VP40. After transfection, VLPs were purified from the cell medium by centrifugation through a sucrose cushion. NCBI-ID: 205488 / Sci species name: Ebola virus sp. / Sci species strain: EBOV-Makona VLP / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Host system | Organism:  Homo sapiens (human) / Recombinant cell: HEK 293T/17 / Recombinant plasmid: pWRG7077 Homo sapiens (human) / Recombinant cell: HEK 293T/17 / Recombinant plasmid: pWRG7077 |
-Supramolecule #1: virus-like particle / antibody complex
| Supramolecule | Name: virus-like particle / antibody complex / type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #3: c2G4 antibody
| Supramolecule | Name: c2G4 antibody / type: complex / ID: 3 / Parent: 1 Details: Whole chimerized human IgG from ZMapp antibody cocktail that binds the base region of Ebola virus glycoproteins |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 10% sucrose-HM (20 mM HEPES, 20 mM MES, 130 mM NaCl) |
| Grid | Model: Quantifoil Multi-A / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 294 K / Instrument: LEICA EM GP Details: Blot for 6 seconds before plunging into liquid ethane (LEICA EM GP).. |
| Details | 8 micrograms of Ebola VLPs were incubated with 12.5 micrograms of c2G4 antibody on ice for ~30-60 minutes before plunge-freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: Gatan, Inc / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 0-4 / Number grids imaged: 2 / Average exposure time: 2.0 sec. / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 23000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER Details: The average resolution of all maps was estimated based on comparison with features observed when X-ray structures are filtered to this resolution. Number subtomograms used: 1998 |
|---|---|
| Extraction | Number tomograms: 26 / Number images used: 2470 / Method: Volumes were picked automatically. |
| Final angle assignment | Type: OTHER |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER Target criteria: Fitting was based on location of antibody escape mutant residues. |
|---|
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)