+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-7822 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of the zebrafish TRPM2 channel in the presence of Ca2+ | |||||||||
 マップデータ マップデータ | Single-particle cryo-EM reconstruction of TRPM2 | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  | |||||||||
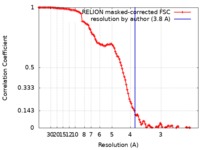
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.8 Å | |||||||||
 データ登録者 データ登録者 | Yin Y / Wu M / Borschel WF / Lander GC / Lee S-Y | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2019 ジャーナル: Nat Commun / 年: 2019タイトル: Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel. 著者: Ying Yin / Mengyu Wu / Allen L Hsu / William F Borschel / Mario J Borgnia / Gabriel C Lander / Seok-Yong Lee /  要旨: The transient receptor potential melastatin 2 (TRPM2) channel plays a key role in redox sensation in many cell types. Channel activation requires binding of both ADP-ribose (ADPR) and Ca. The ...The transient receptor potential melastatin 2 (TRPM2) channel plays a key role in redox sensation in many cell types. Channel activation requires binding of both ADP-ribose (ADPR) and Ca. The recently published TRPM2 structures from Danio rerio in the ligand-free and the ADPR/Ca-bound conditions represent the channel in closed and open states, which uncovered substantial tertiary and quaternary conformational rearrangements. However, it is unclear how these rearrangements are achieved within the tetrameric channel during channel gating. Here we report the cryo-electron microscopy structures of Danio rerio TRPM2 in the absence of ligands, in complex with Ca alone, and with both ADPR and Ca, resolved to ~4.3 Å, ~3.8 Å, and ~4.2 Å, respectively. In contrast to the published results, our studies capture ligand-bound TRPM2 structures in two-fold symmetric intermediate states, offering a glimpse of the structural transitions that bridge the closed and open conformations. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_7822.map.gz emd_7822.map.gz | 113 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-7822-v30.xml emd-7822-v30.xml emd-7822.xml emd-7822.xml | 19.5 KB 19.5 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_7822_fsc.xml emd_7822_fsc.xml | 11.4 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_7822.png emd_7822.png | 246.1 KB | ||
| その他 |  emd_7822_half_map_1.map.gz emd_7822_half_map_1.map.gz emd_7822_half_map_2.map.gz emd_7822_half_map_2.map.gz | 96.4 MB 96.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7822 http://ftp.pdbj.org/pub/emdb/structures/EMD-7822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7822 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_7822_validation.pdf.gz emd_7822_validation.pdf.gz | 776.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_7822_full_validation.pdf.gz emd_7822_full_validation.pdf.gz | 775.6 KB | 表示 | |
| XML形式データ |  emd_7822_validation.xml.gz emd_7822_validation.xml.gz | 18.6 KB | 表示 | |
| CIF形式データ |  emd_7822_validation.cif.gz emd_7822_validation.cif.gz | 24.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7822 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7822 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7822 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7822 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_7822.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_7822.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Single-particle cryo-EM reconstruction of TRPM2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-ハーフマップ: Half 1
| ファイル | emd_7822_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half 1 | ||||||||||||
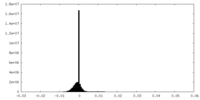
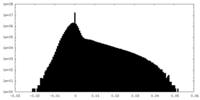
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half 2
| ファイル | emd_7822_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half 2 | ||||||||||||
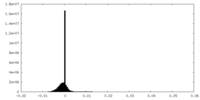
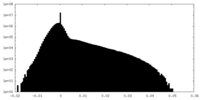
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Transient receptor potential cation channel subfamily M member 2 ...
| 全体 | 名称: Transient receptor potential cation channel subfamily M member 2 (TRPM2) |
|---|---|
| 要素 |
|
-超分子 #1: Transient receptor potential cation channel subfamily M member 2 ...
| 超分子 | 名称: Transient receptor potential cation channel subfamily M member 2 (TRPM2) タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1 |
|---|---|
| 由来(天然) | 生物種:  |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Transient receptor potential cation channel, subfamily M
| 分子 | 名称: Transient receptor potential cation channel, subfamily M タイプ: protein_or_peptide / ID: 1 / コピー数: 4 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 167.608625 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MALGTSGVKI HPNGNSNQLG VQLENVKLTS LFKKLDKRCS LASWIKENIK KKECCFYVED GREGICKCGY PKVQHCDEAI KPEDYMGEQ WDKHRHVRET PTDAFGDISF GGLGQKTGKY VRVSSDTSCE NLYQLMTEQW KLRSPNLLIS VTGGAKNFYI K THLKDKFR ...文字列: MALGTSGVKI HPNGNSNQLG VQLENVKLTS LFKKLDKRCS LASWIKENIK KKECCFYVED GREGICKCGY PKVQHCDEAI KPEDYMGEQ WDKHRHVRET PTDAFGDISF GGLGQKTGKY VRVSSDTSCE NLYQLMTEQW KLRSPNLLIS VTGGAKNFYI K THLKDKFR RGLIKVAQTT GAWILTGGTH AGVMKHVGMA VRDYTLSSGS MEGQIVVIGV APWGVIHNRS TLIHPEGRFP AY YSLDEQG QGRLSCLDIN HTHFLLVDDG TQGHYGVEIE LRARLEKLIS KLSLGNRESG VTIPVVCVVL DGGPGTLNTI YNS MLNHTP CVVLEGSGRL ADVIAHVASV PVSKVTMALI NRLLKRFFMQ EYKNFTELQI IEWTKKIQDI LRMPHLLTVF RIDE DKNYD VDVAILQALL KASRSDEHAG RHCWERQLEL AVAWNRVDIA ESEIFTEESQ WTSSDLHPAM FSALVGDKPE FVRLL LENG VCVREFLERE ETLCELYSHL PSCFFLRKLA KRVQGGKMRR GQEPLPGSRK VCLSHVSEEV RHLLGSFTQP LYIASR YKP TKDDVRLKVP SKGALDLPCS GEEWSADTVW DPGRDLFLWA VVQNNRELAE IGWEQCRDCI AAALAASKIL RKLAQES GE DDSEEATEML ELANHYEKQA IGVFSECHSW DAQRAQKLLI RISPSWGRST CLWLALEAHD KSFIAHSGVQ ALLTQIWC G ELSVDNPHWK VLLCMIFFPL IYTGFLTFRR DEDIQRQAER TEQQKLAMES VFAGQSDGKI KRHLRGFSQK SELKPLNCS SRLMSFLKSP QVKFYWNIAS YFGFLWLFAV VLMIDFQTSP SWRELLLYVW LTSLVCEEIR QLYHDFDGSG FRRKAKMYIK DLWNILDVL SIVLFIAGLI CRLQASDTVF YIGKVILCID FIIFCLRLMA IFSISRTLGP KIIIVRRMML DLFFFMFLLS I WVVAYGVA KQGILIENEE RLNWIIRGAV YEPYITIFGN FPTNIDNTLF DISSCSVNAS DPLKPKCPML NADNTPVFPE WL TIMMLCV YLLFANILLL NLLIAIFNYT FQEVQDNTDT IWKFQRYELI KEYHSRPALP PPFILLSHLI LFIRGVFLRD LPQ RHKNFR QELEQTEEEE LLSWEAYMKD NYLASTRQDE SQSVEHRIHD TAEKVGAMSE LLEREQEMV(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)DEEAPHM FARQLQYPDS TVRRFPVPEE KVSWEVNFSP YQPPVYNQQD SSESDTSALD KHRNPGGR T GIRGKGALNT LGPNHILHPI FTRWRDAEHK VLEFLAVWED AEKRWALLGG PAQPDEPLAQ VLERILGKKL NEKTKTLLK AGEEVYKGYV DDSRNTDNAW VETSIITLHC DKNTPLMADL NHMVESSLSS HQPLQWREVS SDACRCSYQR EALRQIAHHH NTYFSNSLE VLFQGPDYKD DDDKAHHHHH HHHHH |
-分子 #2: CALCIUM ION
| 分子 | 名称: CALCIUM ION / タイプ: ligand / ID: 2 / コピー数: 4 / 式: CA |
|---|---|
| 分子量 | 理論値: 40.078 Da |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.5 mg/mL |
|---|---|
| 緩衝液 | pH: 8 |
| グリッド | モデル: Quantifoil, UltrAuFoil, R1.2/1.3 / 材質: GOLD / メッシュ: 300 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: CONTINUOUS / 前処理 - タイプ: PLASMA CLEANING |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277.15 K / 装置: HOMEMADE PLUNGER |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TALOS ARCTICA |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / デジタル化 - サイズ - 横: 3710 pixel / デジタル化 - サイズ - 縦: 3838 pixel / デジタル化 - サンプリング間隔: 5.0 µm / デジタル化 - 画像ごとのフレーム数: 1-64 / 実像数: 3039 / 平均露光時間: 16.0 sec. / 平均電子線量: 63.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 1.2 µm / 倍率(公称値): 36000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Talos Arctica / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT / 温度因子: 100 |
|---|---|
| 得られたモデル |  PDB-6d73: |
 ムービー
ムービー コントローラー
コントローラー

















 Z
Z Y
Y X
X