[English] 日本語
 Yorodumi
Yorodumi- EMDB-6810: V/A-type ATPase/synthase from Thermus thermophilus, rotational state 1 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6810 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | V/A-type ATPase/synthase from Thermus thermophilus, rotational state 1 | |||||||||
 Map data Map data | The density map of V-type ATP synthase, state 1, from thermophilic bacterium, Thermus thermophilus. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP synthase / proton pump / V-ATPase / Bioenergetics / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton-transporting V-type ATPase, V0 domain / proton-transporting two-sector ATPase complex, catalytic domain / proton-transporting ATP synthase complex / proton motive force-driven plasma membrane ATP synthesis / H+-transporting two-sector ATPase / proton-transporting ATPase activity, rotational mechanism / proton-transporting ATP synthase activity, rotational mechanism / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) | |||||||||
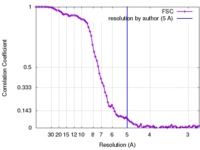
| Method | single particle reconstruction / cryo EM / Resolution: 5.0 Å | |||||||||
 Authors Authors | Nakanishi A / Kishikawa J | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Cryo EM structure of intact rotary H-ATPase/synthase from Thermus thermophilus. Authors: Atsuko Nakanishi / Jun-Ichi Kishikawa / Masatada Tamakoshi / Kaoru Mitsuoka / Ken Yokoyama /  Abstract: Proton translocating rotary ATPases couple ATP hydrolysis/synthesis, which occurs in the soluble domain, with proton flow through the membrane domain via a rotation of the common central rotor ...Proton translocating rotary ATPases couple ATP hydrolysis/synthesis, which occurs in the soluble domain, with proton flow through the membrane domain via a rotation of the common central rotor complex against the surrounding peripheral stator apparatus. Here, we present a large data set of single particle cryo-electron micrograph images of the V/A type H-rotary ATPase from the bacterium Thermus thermophilus, enabling the identification of three rotational states based on the orientation of the rotor subunit. Using masked refinement and classification with signal subtractions, we obtain homogeneous reconstructions for the whole complexes and soluble V domains. These reconstructions are of higher resolution than any EM map of intact rotary ATPase reported previously, providing a detailed molecular basis for how the rotary ATPase maintains structural integrity of the peripheral stator apparatus, and confirming the existence of a clear proton translocation path from both sides of the membrane. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6810.map.gz emd_6810.map.gz | 46.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6810-v30.xml emd-6810-v30.xml emd-6810.xml emd-6810.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6810_fsc.xml emd_6810_fsc.xml | 9.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_6810.png emd_6810.png | 54 KB | ||
| Filedesc metadata |  emd-6810.cif.gz emd-6810.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6810 http://ftp.pdbj.org/pub/emdb/structures/EMD-6810 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6810 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6810 | HTTPS FTP |
-Validation report
| Summary document |  emd_6810_validation.pdf.gz emd_6810_validation.pdf.gz | 606.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6810_full_validation.pdf.gz emd_6810_full_validation.pdf.gz | 606.5 KB | Display | |
| Data in XML |  emd_6810_validation.xml.gz emd_6810_validation.xml.gz | 10.4 KB | Display | |
| Data in CIF |  emd_6810_validation.cif.gz emd_6810_validation.cif.gz | 13.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6810 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6810 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6810 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6810 | HTTPS FTP |
-Related structure data
| Related structure data |  5y5xMC  6811C  6812C  6813C  5y5yC  5y5zC  5y60C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6810.map.gz / Format: CCP4 / Size: 50.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6810.map.gz / Format: CCP4 / Size: 50.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The density map of V-type ATP synthase, state 1, from thermophilic bacterium, Thermus thermophilus. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : V/A-type ATPase/synthase from Thermus thermophilus, rotational st...
+Supramolecule #1: V/A-type ATPase/synthase from Thermus thermophilus, rotational st...
+Macromolecule #1: V-type ATP synthase alpha chain
+Macromolecule #2: V-type ATP synthase beta chain
+Macromolecule #3: V-type ATP synthase subunit D
+Macromolecule #4: V-type ATP synthase subunit F
+Macromolecule #5: V-type ATP synthase, subunit (VAPC-THERM)
+Macromolecule #6: V-type ATP synthase subunit E
+Macromolecule #7: V-type ATP synthase subunit C
+Macromolecule #8: V-type ATP synthase subunit I
+Macromolecule #9: V-type ATP synthase, subunit K
+Macromolecule #10: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.027 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: MOLYBDENUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 1.0 sec. / Average electron dose: 26.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)