[English] 日本語
 Yorodumi
Yorodumi- EMDB-62040: Cryo-EM structure of Arabidopsis thaliana H2A-nucleosome with 147... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Arabidopsis thaliana H2A-nucleosome with 147bp Widom 601 DNA (C2 symmetry) | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleosome / histone / H2A / Arabidopsis / NUCLEAR PROTEIN/DNA / NUCLEAR PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-mediated transformation / chromocenter / thylakoid / response to water deprivation / plasmodesma / plant-type vacuole / plastid / chloroplast / response to bacterium / response to wounding ...DNA-mediated transformation / chromocenter / thylakoid / response to water deprivation / plasmodesma / plant-type vacuole / plastid / chloroplast / response to bacterium / response to wounding / structural constituent of chromatin / nucleosome / peroxisome / protein heterodimerization activity / nucleolus / DNA binding / extracellular region / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.14 Å | |||||||||
 Authors Authors | Wang Y / Dong A | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2025 Journal: Structure / Year: 2025Title: Structural and functional interrelationships of histone H2A with its variants H2A.Z and H2A.W in Arabidopsis. Authors: Youchao Wang / Jiabing Wu / Shuoming Yang / Xiang Li / Jiachen Wang / Qinghe Lv / Xiaoyu Zhu / Guoliang Lu / Jinru Zhang / Wen-Hui Shen / Bing Liu / Jinzhong Lin / Aiwu Dong /   Abstract: Multiple histone H2A variants are known in eukaryotes. However, the functional relationship between H2A and its variants in plants remains largely obscure. Using CRISPR-Cas9 editing, we generated a ...Multiple histone H2A variants are known in eukaryotes. However, the functional relationship between H2A and its variants in plants remains largely obscure. Using CRISPR-Cas9 editing, we generated a mutant lacking four H2A isoforms in Arabidopsis and analyzed the functional and structural relationships between H2A, H2A.Z, and H2A.W. RNA sequencing and phenotype analyses revealed mild changes in gene transcription and plant development in mutants lacking H2A, H2A.Z, or H2A.W compared with the wild-type plants. Chromatin immunoprecipitation sequencing analysis showed that H2A can substitute for both H2A.Z and H2A.W across the genome, including in euchromatin and heterochromatin regions. However, H2A.Z replaced both H2A and H2A.W primarily within the euchromatin regions. By using DNA and histones from Arabidopsis, we constructed nucleosomes containing H2A, H2A.Z, or H2A.W and resolved their cryogenic electron microscopy (cryo-EM) structures at near-atomic resolution. Collectively, the results reveal the structural similarity and functional redundancy of H2A and its variants in Arabidopsis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_62040.map.gz emd_62040.map.gz | 32.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-62040-v30.xml emd-62040-v30.xml emd-62040.xml emd-62040.xml | 24.2 KB 24.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_62040_fsc.xml emd_62040_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_62040.png emd_62040.png | 191.6 KB | ||
| Masks |  emd_62040_msk_1.map emd_62040_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-62040.cif.gz emd-62040.cif.gz | 6.7 KB | ||
| Others |  emd_62040_additional_1.map.gz emd_62040_additional_1.map.gz emd_62040_half_map_1.map.gz emd_62040_half_map_1.map.gz emd_62040_half_map_2.map.gz emd_62040_half_map_2.map.gz | 33 MB 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-62040 http://ftp.pdbj.org/pub/emdb/structures/EMD-62040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62040 | HTTPS FTP |
-Validation report
| Summary document |  emd_62040_validation.pdf.gz emd_62040_validation.pdf.gz | 902.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_62040_full_validation.pdf.gz emd_62040_full_validation.pdf.gz | 902.4 KB | Display | |
| Data in XML |  emd_62040_validation.xml.gz emd_62040_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_62040_validation.cif.gz emd_62040_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62040 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62040 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62040 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62040 | HTTPS FTP |
-Related structure data
| Related structure data |  9k42MC  9k3zC  9k40C  9k41C  9k43C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_62040.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_62040.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.072 Å | ||||||||||||||||||||||||||||||||||||
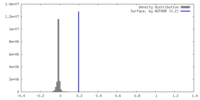
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_62040_msk_1.map emd_62040_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
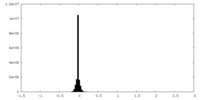
| Density Histograms |
-Additional map: map sharp
| File | emd_62040_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map_sharp | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: map half A
| File | emd_62040_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map_half_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: map half B
| File | emd_62040_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map_half_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Arabidopsis thaliana H2A-nucleosome with 147bp Widom 601 DNA
| Entire | Name: Arabidopsis thaliana H2A-nucleosome with 147bp Widom 601 DNA |
|---|---|
| Components |
|
-Supramolecule #1: Arabidopsis thaliana H2A-nucleosome with 147bp Widom 601 DNA
| Supramolecule | Name: Arabidopsis thaliana H2A-nucleosome with 147bp Widom 601 DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Histone H3.1
| Macromolecule | Name: Histone H3.1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.300968 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RFRPGTVALR EIRKYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VAALQEAAEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.1 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.436467 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK IFLENVIRDA VTYTEHARRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A.6
| Macromolecule | Name: Histone H2A.6 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.680854 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGRGKTLGS GGAKKATSRS SKAGLQFPVG RIARFLKAGK YAERVGAGAP VYLAAVLEYL AAEVLELAGN AARDNKKTRI VPRHIQLAV RNDEELSKLL GDVTIANGGV MPNIHNLLLP KKAGASKPQE D UniProtKB: Histone H2A.6 |
-Macromolecule #4: Histone H2B.1
| Macromolecule | Name: Histone H2B.1 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.439291 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAPRAEKKPA EKKTAAERPV EENKAAEKAP AEKKPKAGKK LPPKEAGDKK KKRSKKNVET YKIYIFKVLK QVHPDIGISS KAMGIMNSF INDIFEKLAQ ESSKLARYNK KPTITSREIQ TAVRLVLPGE LAKHAVSEGT KAVTKFTSS UniProtKB: Histone H2B.1 |
-Macromolecule #5: Widom 601 DNA (147-MER)
| Macromolecule | Name: Widom 601 DNA (147-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.145754 KDa |
| Sequence | String: (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT) ...String: (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT) (DA)(DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT) (DA)(DA) (DA)(DC)(DG)(DC)(DA)(DC)(DG) (DT)(DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT) (DC)(DC)(DC) (DC)(DC)(DG)(DC)(DG)(DT) (DT)(DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC) (DA)(DA)(DG)(DG) (DG)(DG)(DA)(DT)(DT) (DA)(DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT) (DC)(DT)(DC)(DC)(DA) (DG)(DG)(DC)(DA) (DC)(DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC) (DA)(DT)(DC) (DC)(DT)(DG)(DT) |
-Macromolecule #6: Widom 601 DNA (147-MER)
| Macromolecule | Name: Widom 601 DNA (147-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.604047 KDa |
| Sequence | String: (DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT) (DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DG)(DA)(DT) (DT)(DC)(DT) (DC)(DC)(DA)(DG) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Software | Name: EPU |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)