+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focus refinement dmCTPS bound with dATP dUTP dGTP and DON | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | CTPS / filament / inhibitor / substrates / intermediate / LIGASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationInterconversion of nucleotide di- and triphosphates / larval lymph gland hemopoiesis / cytoophidium / CTP synthase (glutamine hydrolysing) / CTP synthase activity / 'de novo' CTP biosynthetic process / pyrimidine nucleobase biosynthetic process / CTP biosynthetic process / ATP binding / identical protein binding ...Interconversion of nucleotide di- and triphosphates / larval lymph gland hemopoiesis / cytoophidium / CTP synthase (glutamine hydrolysing) / CTP synthase activity / 'de novo' CTP biosynthetic process / pyrimidine nucleobase biosynthetic process / CTP biosynthetic process / ATP binding / identical protein binding / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.79 Å | ||||||||||||
 Authors Authors | Guo CJ / Liu JL | ||||||||||||
| Funding support |  China, 1 items China, 1 items
| ||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2024 Journal: J Mol Biol / Year: 2024Title: Structural Basis of Bifunctional CTP/dCTP Synthase. Authors: Chen-Jun Guo / Zherong Zhang / Jia-Li Lu / Jiale Zhong / Yu-Fen Wu / Shu-Ying Guo / Ji-Long Liu /    Abstract: The final step in the de novo synthesis of cytidine 5'-triphosphate (CTP) is catalyzed by CTP synthase (CTPS), which can form cytoophidia in all three domains of life. Recently, we have discovered ...The final step in the de novo synthesis of cytidine 5'-triphosphate (CTP) is catalyzed by CTP synthase (CTPS), which can form cytoophidia in all three domains of life. Recently, we have discovered that CTPS binds to ribonucleotides (NTPs) to form filaments, and have successfully resolved the structures of Drosophila melanogaster CTPS bound with NTPs. Previous biochemical studies have shown that CTPS can bind to deoxyribonucleotides (dNTPs) to produce 2'-deoxycytidine-5'-triphosphate (dCTP). However, the structural basis of CTPS binding to dNTPs is still unclear. In this study, we find that Drosophila CTPS can also form filaments with dNTPs. Using cryo-electron microscopy, we are able to resolve the structure of Drosophila melanogaster CTPS bound to dNTPs with a resolution of up to 2.7 Å. By combining these structural findings with biochemical analysis, we compare the binding and reaction characteristics of NTPs and dNTPs with CTPS. Our results indicate that the same enzyme can act bifunctionally as CTP/dCTP synthase in vitro, and provide a structural basis for these activities. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61009.map.gz emd_61009.map.gz | 32.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61009-v30.xml emd-61009-v30.xml emd-61009.xml emd-61009.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61009_fsc.xml emd_61009_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_61009.png emd_61009.png | 42 KB | ||
| Masks |  emd_61009_msk_1.map emd_61009_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-61009.cif.gz emd-61009.cif.gz | 6 KB | ||
| Others |  emd_61009_additional_1.map.gz emd_61009_additional_1.map.gz emd_61009_half_map_1.map.gz emd_61009_half_map_1.map.gz emd_61009_half_map_2.map.gz emd_61009_half_map_2.map.gz | 935 KB 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61009 http://ftp.pdbj.org/pub/emdb/structures/EMD-61009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61009 | HTTPS FTP |
-Validation report
| Summary document |  emd_61009_validation.pdf.gz emd_61009_validation.pdf.gz | 825.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_61009_full_validation.pdf.gz emd_61009_full_validation.pdf.gz | 825.2 KB | Display | |
| Data in XML |  emd_61009_validation.xml.gz emd_61009_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_61009_validation.cif.gz emd_61009_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61009 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61009 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61009 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61009 | HTTPS FTP |
-Related structure data
| Related structure data |  9iz2MC  9iz1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61009.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61009.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.025 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_61009_msk_1.map emd_61009_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_61009_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61009_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_61009_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : dmCTPS with dATP dUTP dGTP and DON
| Entire | Name: dmCTPS with dATP dUTP dGTP and DON |
|---|---|
| Components |
|
-Supramolecule #1: dmCTPS with dATP dUTP dGTP and DON
| Supramolecule | Name: dmCTPS with dATP dUTP dGTP and DON / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: CTP synthase
| Macromolecule | Name: CTP synthase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: CTP synthase (glutamine hydrolysing) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.443656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYILVTGGV ISGVGKGVIA SSFGTLLKSC GLDVTSIKID PYINIDAGTF SPYEHGEVYV LDDGAEVDLD LGNYERFLDV TLHRDNNIT TGKIYKLVIE KERTGEYLGK TVQVVPHITD AIQEWVERVA QTPVQGSSKP QVCIVELGGT IGDIEGMPFV E AFRQFQFR ...String: MKYILVTGGV ISGVGKGVIA SSFGTLLKSC GLDVTSIKID PYINIDAGTF SPYEHGEVYV LDDGAEVDLD LGNYERFLDV TLHRDNNIT TGKIYKLVIE KERTGEYLGK TVQVVPHITD AIQEWVERVA QTPVQGSSKP QVCIVELGGT IGDIEGMPFV E AFRQFQFR VKRENFCLAH VSLVPLPKAT GEPKTKPTQS SVRELRGCGL SPDLIVCRSE KPIGLEVKEK ISNFCHVGPD QV ICIHDLN SIYHVPLLME QNGVIEYLNE RLQLNIDMSK RTKCLQQWRD LARRTETVRR EVCIAVVGKY TKFTDSYASV VKA LQHAAL AVNRKLELVF IESCLLEEET LHSEPSKYHK EWQKLCDSHG ILVPGGFGSR GMEGKIRACQ WARENQKPLL GICL GLQAA VIEFARNKLG LKDANTTEID PNTANALVID MPEHHTGQLG GTMRLGKRIT VFSDGPSVIR QLYGNPKSVQ ERHRH RYEV NPKYVHLLEE QGMRFVGTDV DKTRMEIIEL SGHPYFVATQ YHPEYLSRPL KPSPPFLGLI LASVDRLNQY IQ UniProtKB: CTP synthase |
-Macromolecule #2: 6-DIAZENYL-5-OXO-L-NORLEUCINE
| Macromolecule | Name: 6-DIAZENYL-5-OXO-L-NORLEUCINE / type: ligand / ID: 2 / Number of copies: 1 / Formula: DON |
|---|---|
| Molecular weight | Theoretical: 173.17 Da |
| Chemical component information | 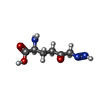 ChemComp-DON: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: 2'-DEOXYADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: DAT |
|---|---|
| Molecular weight | Theoretical: 411.202 Da |
| Chemical component information |  ChemComp-DAT: |
-Macromolecule #5: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: DGT |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-DGT: |
-Macromolecule #6: [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(2-oxidanyl-4-pho...
| Macromolecule | Name: [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(2-oxidanyl-4-phosphonooxy-pyrimidin-1-yl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate type: ligand / ID: 6 / Number of copies: 1 / Formula: 5ZL |
|---|---|
| Molecular weight | Theoretical: 565.129 Da |
| Chemical component information | 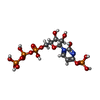 ChemComp-5ZL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 59.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 5.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















































