[English] 日本語
 Yorodumi
Yorodumi- EMDB-60909: The structure of Candida albicans Cdr1 in fluconazole-bound state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of Candida albicans Cdr1 in fluconazole-bound state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporters / Pleiotropic drug resistance / Membrane protein / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationfluconazole transmembrane transporter activity / fluconazole transport / azole transmembrane transport / phosphatidylethanolamine floppase activity / azole transmembrane transporter activity / corticosterone binding / estradiol binding / aminophospholipid flippase activity / phosphatidylserine floppase activity / phosphatidylcholine floppase activity ...fluconazole transmembrane transporter activity / fluconazole transport / azole transmembrane transport / phosphatidylethanolamine floppase activity / azole transmembrane transporter activity / corticosterone binding / estradiol binding / aminophospholipid flippase activity / phosphatidylserine floppase activity / phosphatidylcholine floppase activity / xenobiotic detoxification by transmembrane export across the plasma membrane / phospholipid translocation / response to cycloheximide / xenobiotic transmembrane transporter activity / ABC-type transporter activity / ribonucleoside triphosphate phosphatase activity / extracellular vesicle / response to antibiotic / nucleotide binding / cell surface / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Candida albicans SC5314 (yeast) Candida albicans SC5314 (yeast) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Peng Y / Sun H / Yan ZF | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms. Authors: Ying Peng / Yan Lu / Hui Sun / Jinying Ma / Xiaomei Li / Xiaodan Han / Zhixiong Fang / Junming Tan / Yingchen Qiu / Tingting Qu / Meng Yin / Zhaofeng Yan /  Abstract: In Candida albicans, Cdr1 pumps azole drugs out of the cells to reduce intracellular accumulation at detrimental concentrations, leading to azole-drug resistance. Milbemycin oxime, a veterinary anti- ...In Candida albicans, Cdr1 pumps azole drugs out of the cells to reduce intracellular accumulation at detrimental concentrations, leading to azole-drug resistance. Milbemycin oxime, a veterinary anti-parasitic drug, strongly and specifically inhibits Cdr1. However, how Cdr1 recognizes and exports azole drugs, and how milbemycin oxime inhibits Cdr1 remain unclear. Here, we report three cryo-EM structures of Cdr1 in distinct states: the apo state (Cdr1), fluconazole-bound state (Cdr1), and milbemycin oxime-inhibited state (Cdr1). Both the fluconazole substrate and the milbemycin oxime inhibitor are primarily recognized within the central cavity of Cdr1 through hydrophobic interactions. The fluconazole is suggested to be exported from the binding site into the environment through a lateral pathway driven by TM2, TM5, TM8 and TM11. Our findings uncover the inhibitory mechanism of milbemycin oxime, which inhibits Cdr1 through competition, hindering export, and obstructing substrate entry. These discoveries advance our understanding of Cdr1-mediated azole resistance in C. albicans and provide the foundation for the development of innovative antifungal drugs targeting Cdr1 to combat azole-drug resistance. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60909.map.gz emd_60909.map.gz | 12.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60909-v30.xml emd-60909-v30.xml emd-60909.xml emd-60909.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60909_fsc.xml emd_60909_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_60909.png emd_60909.png | 105.5 KB | ||
| Filedesc metadata |  emd-60909.cif.gz emd-60909.cif.gz | 6.5 KB | ||
| Others |  emd_60909_half_map_1.map.gz emd_60909_half_map_1.map.gz emd_60909_half_map_2.map.gz emd_60909_half_map_2.map.gz | 80.9 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60909 http://ftp.pdbj.org/pub/emdb/structures/EMD-60909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60909 | HTTPS FTP |
-Validation report
| Summary document |  emd_60909_validation.pdf.gz emd_60909_validation.pdf.gz | 738.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60909_full_validation.pdf.gz emd_60909_full_validation.pdf.gz | 737.6 KB | Display | |
| Data in XML |  emd_60909_validation.xml.gz emd_60909_validation.xml.gz | 17.7 KB | Display | |
| Data in CIF |  emd_60909_validation.cif.gz emd_60909_validation.cif.gz | 22.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60909 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60909 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60909 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60909 | HTTPS FTP |
-Related structure data
| Related structure data |  9iulMC  9iukC  9iumC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60909.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60909.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60909_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60909_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The Cryo-EM structure of Candida albicans Cdr1 in fluconazole-bou...
| Entire | Name: The Cryo-EM structure of Candida albicans Cdr1 in fluconazole-bound state |
|---|---|
| Components |
|
-Supramolecule #1: The Cryo-EM structure of Candida albicans Cdr1 in fluconazole-bou...
| Supramolecule | Name: The Cryo-EM structure of Candida albicans Cdr1 in fluconazole-bound state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Candida albicans SC5314 (yeast) Candida albicans SC5314 (yeast) |
-Macromolecule #1: Pleiotropic ABC efflux transporter of multiple drugs CDR1
| Macromolecule | Name: Pleiotropic ABC efflux transporter of multiple drugs CDR1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candida albicans SC5314 (yeast) Candida albicans SC5314 (yeast) |
| Molecular weight | Theoretical: 170.159656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDSKMSSQD ESKLEKAISQ DSSSENHSIN EYHGFDAHTS ENIQNLARTF THDSFKDDSS AGLLKYLTHM SEVPGVNPYE HEEINNDQL NPDSENFNAK FWVKNLRKLF ESDPEYYKPS KLGIGYRNLR AYGVANDSDY QPTVTNALWK LATEGFRHFQ K DDDSRYFD ...String: MSDSKMSSQD ESKLEKAISQ DSSSENHSIN EYHGFDAHTS ENIQNLARTF THDSFKDDSS AGLLKYLTHM SEVPGVNPYE HEEINNDQL NPDSENFNAK FWVKNLRKLF ESDPEYYKPS KLGIGYRNLR AYGVANDSDY QPTVTNALWK LATEGFRHFQ K DDDSRYFD ILKSMDAIMR PGELTVVLGR PGAGCSTLLK TIAVNTYGFH IGKESQITYD GLSPHDIERH YRGDVIYSAE TD VHFPHLS VGDTLEFAAR LRTPQNRGEG IDRETYAKHM ASVYMATYGL SHTRNTNVGN DFVRGVSGGE RKRVSIAEAS LSG ANIQCW DNATRGLDSA TALEFIRALK TSAVILDTTP LIAIYQCSQD AYDLFDKVVV LYEGYQIFFG KATKAKEYFE KMGW KCPQR QTTADFLTSL TNPAEREPLP GYEDKVPRTA QEFETYWKNS PEYAELTKEI DEYFVECERS NTRETYRESH VAKQS NNTR PASPYTVSFF MQVRYGVARN FLRMKGDPSI PIFSVFGQLV MGLILSSVFY NLSQTTGSFY YRGAAMFFAV LFNAFS SLL EIMSLFEARP IVEKHKKYAL YRPSADALAS IISELPVKLA MSMSFNFVFY FMVNFRRNPG RFFFYWLMCI WCTFVMS HL FRSIGAVSTS ISGAMTPATV LLLAMVIYTG FVIPTPSMLG WSRWINYINP VGYVFESLMV NEFHGREFQC AQYVPSGP G YENISRSNQV CTAVGSVPGN EMVSGTNYLA GAYQYYNSHK WRNLGITIGF AVFFLAIYIA LTEFNKGAMQ KGEIVLFLK GSLKKHKRKT AASNKGDIEA GPVAGKLDYQ DEAEAVNNEK FTEKGSTGSV DFPENREIFF WRDLTYQVKI KKEDRVILDH VDGWVKPGQ ITALMGASGA GKTTLLNCLS ERVTTGIITD GERLVNGHAL DSSFQRSIGY VQQQDVHLET TTVREALQFS A YLRQSNKI SKKEKDDYVD YVIDLLEMTD YADALVGVAG EGLNVEQRKR LTIGVELVAK PKLLLFLDEP TSGLDSQTAW SI CKLMRKL ADHGQAILCT IHQPSALIMA EFDRLLFLQK GGRTAYFGEL GENCQTMINY FEKYGADPCP KEANPAEWML QVV GAAPGS HAKQDYFEVW RNSSEYQAVR EEINRMEAEL SKLPRDNDPE ALLKYAAPLW KQYLLVSWRT IVQDWRSPGY IYSK IFLVV SAALFNGFSF FKAKNNMQGL QNQMFSVFMF FIPFNTLVQQ MLPYFVKQRD VYEVREAPSR TFSWFAFIAG QITSE IPYQ VAVGTIAFFC WYYPLGLYNN ATPTDSVNPR GVLMWMLVTA FYVYTATMGQ LCMSFSELAD NAANLATLLF TMCLNF CGV LAGPDVLPGF WIFMYRCNPF TYLVQAMLST GLANTFVKCA EREYVSVKPP NGESCSTYLD PYIKFAGGYF ETRNDGS CA FCQMSSTNTF LKSVNSLYSE RWRNFGIFIA FIAINIILTV IFYWLARVPK GNREKKNKK UniProtKB: Pleiotropic ABC efflux transporter of multiple drugs CDR1 |
-Macromolecule #2: Pip2(20:4/18:0)
| Macromolecule | Name: Pip2(20:4/18:0) / type: ligand / ID: 2 / Number of copies: 1 / Formula: A1L26 |
|---|---|
| Molecular weight | Theoretical: 1.047088 KDa |
-Macromolecule #3: 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL
| Macromolecule | Name: 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL type: ligand / ID: 3 / Number of copies: 1 / Formula: TPF |
|---|---|
| Molecular weight | Theoretical: 306.271 Da |
| Chemical component information | 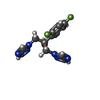 ChemComp-TPF: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.5 µm |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)