+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of MxaF/MxaJ/PQQ complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MxaF/MxaJ/PQQ / PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationmethanol catabolic process / Oxidoreductases; Acting on the CH-OH group of donors; With a cytochrome as acceptor / oxidoreductase activity, acting on CH-OH group of donors / outer membrane-bounded periplasmic space / periplasmic space / calcium ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Methylorubrum extorquens (bacteria) Methylorubrum extorquens (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.81 Å | |||||||||
 Authors Authors | Sun JQ / Gao F | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of MxaF/MxaJ complex Authors: Sun JQ / Gao F | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60840.map.gz emd_60840.map.gz | 89 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60840-v30.xml emd-60840-v30.xml emd-60840.xml emd-60840.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60840.png emd_60840.png | 49.3 KB | ||
| Filedesc metadata |  emd-60840.cif.gz emd-60840.cif.gz | 5.8 KB | ||
| Others |  emd_60840_half_map_1.map.gz emd_60840_half_map_1.map.gz emd_60840_half_map_2.map.gz emd_60840_half_map_2.map.gz | 165.4 MB 165.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60840 http://ftp.pdbj.org/pub/emdb/structures/EMD-60840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60840 | HTTPS FTP |
-Validation report
| Summary document |  emd_60840_validation.pdf.gz emd_60840_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60840_full_validation.pdf.gz emd_60840_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_60840_validation.xml.gz emd_60840_validation.xml.gz | 15 KB | Display | |
| Data in CIF |  emd_60840_validation.cif.gz emd_60840_validation.cif.gz | 17.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60840 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60840 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60840 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60840 | HTTPS FTP |
-Related structure data
| Related structure data |  9isoMC  9ismC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60840.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60840.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_60840_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_60840_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MxaF/MxaJ/PQQ complex
| Entire | Name: MxaF/MxaJ/PQQ complex |
|---|---|
| Components |
|
-Supramolecule #1: MxaF/MxaJ/PQQ complex
| Supramolecule | Name: MxaF/MxaJ/PQQ complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Methylorubrum extorquens (bacteria) Methylorubrum extorquens (bacteria) |
-Macromolecule #1: Methanol dehydrogenase, alpha subunit
| Macromolecule | Name: Methanol dehydrogenase, alpha subunit / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: Oxidoreductases; Acting on the CH-OH group of donors; With a cytochrome as acceptor |
|---|---|
| Source (natural) | Organism:  Methylorubrum extorquens (bacteria) Methylorubrum extorquens (bacteria) |
| Molecular weight | Theoretical: 69.335836 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSRFVTSVSA LAMLALAPAA LSSGAYANDK LVELSKSDDN WVMPGKNYDS NNFSDLKQIN KGNVKQLRPA WTFSTGLLNG HEGAPLVVD GKMYIHTSFP NNTFALGLDD PGTILWQDKP KQNPAARAVA CCDLVNRGLA YWPGDGKTPA LILKTQLDGN V AALNAETG ...String: MSRFVTSVSA LAMLALAPAA LSSGAYANDK LVELSKSDDN WVMPGKNYDS NNFSDLKQIN KGNVKQLRPA WTFSTGLLNG HEGAPLVVD GKMYIHTSFP NNTFALGLDD PGTILWQDKP KQNPAARAVA CCDLVNRGLA YWPGDGKTPA LILKTQLDGN V AALNAETG ETVWKVENSD IKVGSTLTIA PYVVKDKVII GSSGAELGVR GYLTAYDVKT GEQVWRAYAT GPDKDLLLAS DF NIKNPHY GQKGLGTGTW EGDAWKIGGG TNWGWYAYDP GTNLIYFGTG NPAPWNETMR PGDNKWTMTI FGRDADTGEA KFG YQKTPH DEWDYAGVNV MMLSEQKDKD GKARKLLTHP DRNGIVYTLD RTDGALVSAN KLDDTVNVFK SVDLKTGQPV RDPE YGTRM DHLAKDICPS AMGYHNQGHD SYDPKRELFF MGINHICMDW EPFMLPYRAG QFFVGATLNM YPGPKGDRQN YEGLG QIKA YNAITGDYKW EKMERFAVWG GTMATAGDLV FYGTLDGYLK ARDSDTGDLL WKFKIPSGAI GYPMTYTHKG TQYVAI YYG VGGWPGVGLV FDLADPTAGL GAVGAFKKLA NYTQMGGGVV VFSLDGKGPY DDPNVGEWKS AAKHHHHHH UniProtKB: Methanol dehydrogenase, alpha subunit |
-Macromolecule #2: Methanol oxidation protein MxaJ
| Macromolecule | Name: Methanol oxidation protein MxaJ / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Methylorubrum extorquens (bacteria) Methylorubrum extorquens (bacteria) |
| Molecular weight | Theoretical: 32.468154 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLVNGRRRT AASVVALTAA LTALAALCAP AQAQDTKATS KAAEAAKPDA GTLRVCAAEQ PPLSMKDGSG LENRIATTVA EAMGRKAQF VWLGKPAIYL VRDGLEKKTC DVVIGLDADD PRVLTSKPYY RSGYVFLTRA DKDLDIKSWS DPRLKEVSHM V VGFGTPGE ...String: MSLVNGRRRT AASVVALTAA LTALAALCAP AQAQDTKATS KAAEAAKPDA GTLRVCAAEQ PPLSMKDGSG LENRIATTVA EAMGRKAQF VWLGKPAIYL VRDGLEKKTC DVVIGLDADD PRVLTSKPYY RSGYVFLTRA DKDLDIKSWS DPRLKEVSHM V VGFGTPGE AMLKDIGRYE EDMAYLYSLV NFRAPRNQYT QIDPARMVSE VATGKAEVGV AFGPDVARYV RDSSTKLRMT PV PDDTQAS DGRKMPQSFD QAMGVRKDDT ALKAEIDAAL EKAKPKIEAI LKEEGVPVLP VSN UniProtKB: Methanol oxidation protein MxaJ |
-Macromolecule #3: PYRROLOQUINOLINE QUINONE
| Macromolecule | Name: PYRROLOQUINOLINE QUINONE / type: ligand / ID: 3 / Number of copies: 2 / Formula: PQQ |
|---|---|
| Molecular weight | Theoretical: 330.206 Da |
| Chemical component information | 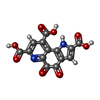 ChemComp-PQQ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































