+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Orf virus scaffolding protein Orfv075 | |||||||||
 Map data Map data | 3D reconstruction of Orfv075 (sharpened) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Scaffold protein / Capsid protein / Poxvirus / VIRAL PROTEIN | |||||||||
| Function / homology | Poxvirus rifampicin-resistance / Poxvirus rifampicin resistance protein / response to antibiotic / 62 kDa protein Function and homology information Function and homology information | |||||||||
| Biological species |  Orf virus (strain NZ2) Orf virus (strain NZ2) | |||||||||
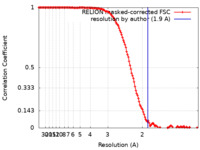
| Method | single particle reconstruction / cryo EM / Resolution: 1.9 Å | |||||||||
 Authors Authors | Hyun J / Kim S / Ko S / Kim M / Jang Y | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: Biochem Biophys Res Commun / Year: 2024 Journal: Biochem Biophys Res Commun / Year: 2024Title: Cryo-EM structure of orf virus scaffolding protein orfv075. Authors: Seungmi Kim / Sumin Ko / Minjae Kim / Yeontae Jang / Jaekyung Hyun /  Abstract: Capsid-like poxvirus scaffold proteins self-assemble into semi-regular lattice that govern the formation of spherical immature virus particles. The scaffolding is a critical step in virus ...Capsid-like poxvirus scaffold proteins self-assemble into semi-regular lattice that govern the formation of spherical immature virus particles. The scaffolding is a critical step in virus morphogenesis as exemplified by the drug rifampicin that impairs the recruitment of scaffold onto the viral membrane in vaccinia virus (VACV). Here we report cryo-electron microscopy structure of scaffolding protein Orfv075 of orf virus (ORFV) that causes smallpox-like diseases in sheep, goats and occasionally humans via zoonotic infection. We demonstrate that the regions that are involved in intertrimeric interactions for scaffold assembly are largely conserved in comparison to its VACV orthologue protein D13 whose intermediate assembly structures have been previously characterized. By contrast, less conserved regions are located away from these interfaces, indicating both viruses share similar assembly mechanisms. We also show that the phenylalanine-rich binding site of rifampicin in D13 is conserved in Orfv075, and molecular docking simulation confirms similar binding modes. Our study provides structural basis of scaffolding protein as a target for anti-poxvirus treatment across wide range of poxvirus genera. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60655.map.gz emd_60655.map.gz | 203.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60655-v30.xml emd-60655-v30.xml emd-60655.xml emd-60655.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60655_fsc.xml emd_60655_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_60655.png emd_60655.png | 180.7 KB | ||
| Masks |  emd_60655_msk_1.map emd_60655_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-60655.cif.gz emd-60655.cif.gz | 7 KB | ||
| Others |  emd_60655_additional_1.map.gz emd_60655_additional_1.map.gz emd_60655_half_map_1.map.gz emd_60655_half_map_1.map.gz emd_60655_half_map_2.map.gz emd_60655_half_map_2.map.gz | 108.7 MB 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60655 http://ftp.pdbj.org/pub/emdb/structures/EMD-60655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60655 | HTTPS FTP |
-Validation report
| Summary document |  emd_60655_validation.pdf.gz emd_60655_validation.pdf.gz | 1017.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60655_full_validation.pdf.gz emd_60655_full_validation.pdf.gz | 1017.2 KB | Display | |
| Data in XML |  emd_60655_validation.xml.gz emd_60655_validation.xml.gz | 20.6 KB | Display | |
| Data in CIF |  emd_60655_validation.cif.gz emd_60655_validation.cif.gz | 26.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60655 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60655 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60655 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60655 | HTTPS FTP |
-Related structure data
| Related structure data |  9ikcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60655.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60655.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of Orfv075 (sharpened) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
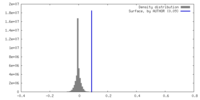
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_60655_msk_1.map emd_60655_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
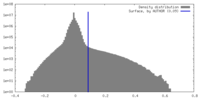
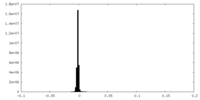
| Density Histograms |
-Additional map: 3D reconstruction of Orfv075 (unsharpened)
| File | emd_60655_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of Orfv075 (unsharpened) | ||||||||||||
| Projections & Slices |
| ||||||||||||
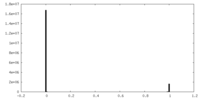
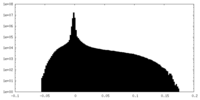
| Density Histograms |
-Half map: First halfmap of Orfv075 3D reconstruction
| File | emd_60655_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First halfmap of Orfv075 3D reconstruction | ||||||||||||
| Projections & Slices |
| ||||||||||||
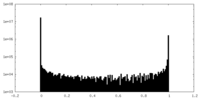
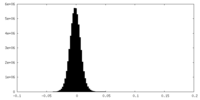
| Density Histograms |
-Half map: Second halfmap of Orfv075 3D reconstruction
| File | emd_60655_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second halfmap of Orfv075 3D reconstruction | ||||||||||||
| Projections & Slices |
| ||||||||||||
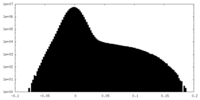
| Density Histograms |
- Sample components
Sample components
-Entire : Orfv075 trimer
| Entire | Name: Orfv075 trimer |
|---|---|
| Components |
|
-Supramolecule #1: Orfv075 trimer
| Supramolecule | Name: Orfv075 trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Orf virus (strain NZ2) Orf virus (strain NZ2) |
| Molecular weight | Theoretical: 190 KDa |
-Macromolecule #1: 62 kDa protein
| Macromolecule | Name: 62 kDa protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Orf virus (strain NZ2) Orf virus (strain NZ2) |
| Molecular weight | Theoretical: 63.652172 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYYHHHHHH DYDIPTTENL YFQGAMNNSV VSLVGGDDAV RRASVFATDH RAPTVYMPQY ITTQGVVDTT SDAVTVTFEI RDKYISAMN NFVLSVDLPE IKGVGKMCYV PYIAYKLIRH VAVNSAADTI WETSGEELFD SCLDNERVME LSGFSRELND L STGSSPND ...String: MSYYHHHHHH DYDIPTTENL YFQGAMNNSV VSLVGGDDAV RRASVFATDH RAPTVYMPQY ITTQGVVDTT SDAVTVTFEI RDKYISAMN NFVLSVDLPE IKGVGKMCYV PYIAYKLIRH VAVNSAADTI WETSGEELFD SCLDNERVME LSGFSRELND L STGSSPND VIKEAACVHA YIKTPFDADK TFSTLKLSDS KVTVTVTLNP VACVMVYDET FDAAKLAKEF PYSMELSFIG YM VKNLCPR PAFIEMPRRR VEQINHTTAV ITDVHACTSL SVYMKPVLSD ANNRFISYPG FQQSEGDFVM AFVERLLEDM VIV SNCYPE GFPETAEIVE VPPSGVVSIQ DTDVFVRIDD VPVGMRVFLH TNILVFATRK NSVVYNMSKK FSAITGAYSR ATSR IRFTT AIHSVNIGDA SVPVGVWTCQ RNVYNGDNRS PEARAKDLFV ADPFLKGVDF KNKIDVIARM DVRFGNEVLY SENSA VSRV FGEILGKTPG VRTLQFNFTP STFFSPTALN SNVSRGKDKL AVRVTTAHME AHNPLMYVPR QMVVVCNEVY RLSYDA GIV AEKVTAQ UniProtKB: 62 kDa protein |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 150 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.13 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 93.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 10975 / Average exposure time: 2.6 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 76923 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 1-547 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Structure of orthologue protein D13 (PDB 7VFD) was fitted into the density map of orfv075 using UCSF Chimera. The fitted structure was modified in Coot based on sequence alignment between D13 and Orfv075. The resulting Orfv075 model was refined using PHENIX. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-9ikc: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)