[English] 日本語
 Yorodumi
Yorodumi- EMDB-60647: Cryo-EM structure of the human P2X3 receptor-compound 26a complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | P2X3 receptor / compound 26a / Cryo-EM / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationPlatelet homeostasis / extracellularly ATP-gated monoatomic cation channel activity / purinergic nucleotide receptor activity / peristalsis / Elevation of cytosolic Ca2+ levels / neuromuscular synaptic transmission / urinary bladder smooth muscle contraction / response to carbohydrate / inorganic cation transmembrane transport / cellular response to ATP ...Platelet homeostasis / extracellularly ATP-gated monoatomic cation channel activity / purinergic nucleotide receptor activity / peristalsis / Elevation of cytosolic Ca2+ levels / neuromuscular synaptic transmission / urinary bladder smooth muscle contraction / response to carbohydrate / inorganic cation transmembrane transport / cellular response to ATP / protein homotrimerization / behavioral response to pain / positive regulation of calcium ion transport into cytosol / response to mechanical stimulus / positive regulation of calcium-mediated signaling / response to cold / hippocampal mossy fiber to CA3 synapse / establishment of localization in cell / calcium ion transmembrane transport / regulation of synaptic plasticity / Schaffer collateral - CA1 synapse / sensory perception of taste / response to heat / response to hypoxia / receptor complex / postsynapse / axon / signal transduction / ATP binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.61 Å | |||||||||
 Authors Authors | Kim S / Kim GR / Kim YO / Han X / Nagel J / Kim J / Song DI / Muller CE / Yoon MH / Jin MS / Kim YC | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: J Med Chem / Year: 2024 Journal: J Med Chem / Year: 2024Title: Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy. Authors: Ga-Ram Kim / Subin Kim / Yeo-Ok Kim / Xuehao Han / Jessica Nagel / Jihyun Kim / Dahin Irene Song / Christa E Müller / Myung-Ha Yoon / Mi Sun Jin / Yong-Chul Kim /    Abstract: The P2X3 receptor (P2X3R), an ATP-gated cation channel predominantly expressed in C- and Aδ-primary afferent neurons, has been proposed as a drug target for neurological inflammatory diseases, e.g., ...The P2X3 receptor (P2X3R), an ATP-gated cation channel predominantly expressed in C- and Aδ-primary afferent neurons, has been proposed as a drug target for neurological inflammatory diseases, e.g., neuropathic pain, and chronic cough. Aiming to develop novel, selective P2X3R antagonists, tetrazolopyrimidine-based hit compound was optimized through structure-activity relationship studies by modifying the tetrazole core as well as side chain substituents. The optimized antagonist , featuring a cyclopropane-substituted triazolopyrimidine core, displayed potent P2X3R-antagonistic activity (IC = 54.9 nM), 20-fold selectivity versus the heteromeric P2X2/3R, and high selectivity versus other P2XR subtypes. Noncompetitive P2X3R blockade was experimentally confirmed by calcium influx assays. Cryo-electron microscopy revealed that stabilizes the P2X3R in its desensitized state, acting as a molecular barrier to prevent ions from accessing the central pore. In vivo studies in a rat neuropathic pain model (spinal nerve ligation) showed dose-dependent antiallodynic effects of , thus presenting a novel, promising lead structure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60647.map.gz emd_60647.map.gz | 30.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60647-v30.xml emd-60647-v30.xml emd-60647.xml emd-60647.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60647.png emd_60647.png | 151.8 KB | ||
| Filedesc metadata |  emd-60647.cif.gz emd-60647.cif.gz | 6.4 KB | ||
| Others |  emd_60647_half_map_1.map.gz emd_60647_half_map_1.map.gz emd_60647_half_map_2.map.gz emd_60647_half_map_2.map.gz | 55.4 MB 55.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60647 http://ftp.pdbj.org/pub/emdb/structures/EMD-60647 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60647 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60647 | HTTPS FTP |
-Validation report
| Summary document |  emd_60647_validation.pdf.gz emd_60647_validation.pdf.gz | 827.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60647_full_validation.pdf.gz emd_60647_full_validation.pdf.gz | 827.1 KB | Display | |
| Data in XML |  emd_60647_validation.xml.gz emd_60647_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_60647_validation.cif.gz emd_60647_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60647 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60647 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60647 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60647 | HTTPS FTP |
-Related structure data
| Related structure data |  9ik1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60647.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60647.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60647_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60647_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human P2X3 receptor-compound 26a complex
| Entire | Name: Human P2X3 receptor-compound 26a complex |
|---|---|
| Components |
|
-Supramolecule #1: Human P2X3 receptor-compound 26a complex
| Supramolecule | Name: Human P2X3 receptor-compound 26a complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: P2X purinoceptor 3
| Macromolecule | Name: P2X purinoceptor 3 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.37352 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DFFTYETPKV IVVKSWTIGI INRVVQLLII SYFVGWVFLH EKAYQVRDTA IESSVVTKVK GSGLYANRVM DVSDYVTPPQ GTSVFVIIT KMIVTENQMQ GFCPESEEKY RCVSDSQCGP ERLPGGGILT GRCVNYSSVL RTCEIQGWCP TEVDTVETPI M MEAENFTI ...String: DFFTYETPKV IVVKSWTIGI INRVVQLLII SYFVGWVFLH EKAYQVRDTA IESSVVTKVK GSGLYANRVM DVSDYVTPPQ GTSVFVIIT KMIVTENQMQ GFCPESEEKY RCVSDSQCGP ERLPGGGILT GRCVNYSSVL RTCEIQGWCP TEVDTVETPI M MEAENFTI FIKNSIRFPL FNFEKGNLLP NLTARDMKTC RFHPDKDPFC PILRVGDVVK FAGQDFAKLA RTGGVLGIKI GW VCDLDKA WDQCIPKYSF TRLDSVSEKS SVSPGYNFRF AKYYKMENGS EYRTLLKAFG IRFDVLVYGN AGKFNIIPTI ISS VAAFTS VGVGTVLCDI ILLNFLKGAD QYKAKKFEEV NET UniProtKB: P2X purinoceptor 3 |
-Macromolecule #2: 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4...
| Macromolecule | Name: 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide type: ligand / ID: 2 / Number of copies: 3 / Formula: A1L2M |
|---|---|
| Molecular weight | Theoretical: 456.543 Da |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 3 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


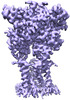
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































