+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fusobacterium nucleatum adhesin CbpF | |||||||||
 Map data Map data | Main map file, sharpened with DeepEMhancer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial pathogenesis / adhesin / type V secretion system / trimeric autotransporter / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Fusobacterium nucleatum (bacteria) Fusobacterium nucleatum (bacteria) | |||||||||
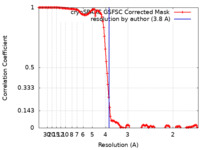
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Roderer D / Marongiu GL / Fink U | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Structural basis for immune cell binding of via the trimeric autotransporter adhesin CbpF. Authors: Gian Luca Marongiu / Uwe Fink / Felix Schöpf / Andreas Oder / Jens Peter von Kries / Daniel Roderer /  Abstract: (Fn), a commensal in the human oral cavity, is overrepresented in the colon microbiota of colorectal cancer (CRC) patients and is linked to tumor chemoresistance, metastasis, and a poor therapeutic ... (Fn), a commensal in the human oral cavity, is overrepresented in the colon microbiota of colorectal cancer (CRC) patients and is linked to tumor chemoresistance, metastasis, and a poor therapeutic prognosis. Fn produces numerous adhesins that mediate tumor colonization and downregulation of the host's antitumor immune response. One of these, the trimeric autotransporter adhesin (TAA) CEACAM binding protein of (CbpF), targets CEACAM1 on T-cells and has been associated with immune evasion of Fn-colonized tumors. Whereas the role of CEACAM1 in homophilic and heterophilic cell interactions and immune evasion is well described, the mechanistic details of its interaction with fusobacterial CbpF remain unknown due to the lack of a high-resolution structure of the adhesin-receptor complex. Here, we present two structures of CbpF alone and in complex with CEACAM1, obtained by cryogenic electron microscopy and single particle analysis. They reveal that CbpF forms a stable homotrimeric complex whose N-terminal part of the extracellular domain comprises a 64 Å long β roll domain with a unique lateral loop extension. CEACAM1 binds to this loop with high affinity via its N-terminal IgV-like domain with a nanomolar dissociation constant as determined by surface plasmon resonance. This study provides a comprehensive structural description of a fusobacterial TAA, illustrates a yet undescribed CEACAM1 binding mode, and paves the way for rational drug design targeting Fn in CRC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51346.map.gz emd_51346.map.gz | 188.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51346-v30.xml emd-51346-v30.xml emd-51346.xml emd-51346.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_51346_fsc.xml emd_51346_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_51346.png emd_51346.png | 91.5 KB | ||
| Masks |  emd_51346_msk_1.map emd_51346_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-51346.cif.gz emd-51346.cif.gz | 6.7 KB | ||
| Others |  emd_51346_additional_1.map.gz emd_51346_additional_1.map.gz emd_51346_half_map_1.map.gz emd_51346_half_map_1.map.gz emd_51346_half_map_2.map.gz emd_51346_half_map_2.map.gz | 107.3 MB 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51346 http://ftp.pdbj.org/pub/emdb/structures/EMD-51346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51346 | HTTPS FTP |
-Validation report
| Summary document |  emd_51346_validation.pdf.gz emd_51346_validation.pdf.gz | 757.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_51346_full_validation.pdf.gz emd_51346_full_validation.pdf.gz | 757.1 KB | Display | |
| Data in XML |  emd_51346_validation.xml.gz emd_51346_validation.xml.gz | 21.5 KB | Display | |
| Data in CIF |  emd_51346_validation.cif.gz emd_51346_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51346 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51346 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51346 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51346 | HTTPS FTP |
-Related structure data
| Related structure data |  9gh4MC  9gh5C  9gh6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_51346.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51346.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map file, sharpened with DeepEMhancer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_51346_msk_1.map emd_51346_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Non-sharpened full map
| File | emd_51346_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-sharpened full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_51346_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_51346_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homotrimeric complex of CbpF, residues 25-274 resolved
| Entire | Name: Homotrimeric complex of CbpF, residues 25-274 resolved |
|---|---|
| Components |
|
-Supramolecule #1: Homotrimeric complex of CbpF, residues 25-274 resolved
| Supramolecule | Name: Homotrimeric complex of CbpF, residues 25-274 resolved type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: nanodisc-embedded CbpF |
|---|---|
| Source (natural) | Organism:  Fusobacterium nucleatum (bacteria) Fusobacterium nucleatum (bacteria) |
| Molecular weight | Theoretical: 160 KDa |
-Macromolecule #1: Cell surface protein
| Macromolecule | Name: Cell surface protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Fusobacterium nucleatum (bacteria) Fusobacterium nucleatum (bacteria) |
| Molecular weight | Theoretical: 51.827051 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKTAIAIAV ALAGFATVAQ ASAAPVIKAG TATDSTEAGV DNVANGVKSS AFGYDNKAIE KESSAFGTGN RATGEFSSAF GFHNIASKI HSSAFGSNNA ADGVNSSAFG FKNTVSGFNS SAFGSQYQVT GNFSGAFGMG EFNGQYQYKN EGNNSYMIGN K NKIASGSD ...String: MKKTAIAIAV ALAGFATVAQ ASAAPVIKAG TATDSTEAGV DNVANGVKSS AFGYDNKAIE KESSAFGTGN RATGEFSSAF GFHNIASKI HSSAFGSNNA ADGVNSSAFG FKNTVSGFNS SAFGSQYQVT GNFSGAFGMG EFNGQYQYKN EGNNSYMIGN K NKIASGSD DNFILGNNVH IGGGINNSVA LGNNSTVSAS NTVSVGSSTL KRKIVNVGDG AISANSSDAV TGRQLYSGNG ID TAAWQNK LNVTRKNDYK DANDIDVNKW KAKLGVGSGG GGGAPVDAYT KSEADNKFAN KTDLNDYTKK DDYKDANGID VDK WKAKLG TGAGTADIEN LRNEVNEKID DVKDEVRTVG SLSAALAGLH PMQYDPKAPV QVMAALGHYR DKQSVAVGAS YYFN DRFMM STGIALSGEK RTKTMANVGF TLKLGKGSGV TYDETPQYVV QNEVKRLTVE NQELKERVRN LEEKLNMLLK NKRSS AWSH PQFEK UniProtKB: Cell surface protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 285 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 79.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-9gh4: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)