[English] 日本語
 Yorodumi
Yorodumi- EMDB-50415: Subtomogram average of 80S ribosomes in S. cerevisiae under acute... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of 80S ribosomes in S. cerevisiae under acute glucose starvation | |||||||||
 Map data Map data | Subtomogram average of 80S ribosomes in S. cerevisiae under acute glucose starvation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Globular protein / translation / RIBOSOME | |||||||||
| Biological species |  | |||||||||
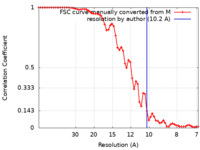
| Method | subtomogram averaging / cryo EM / Resolution: 10.2 Å | |||||||||
 Authors Authors | Spindler MC / Mahamid J | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Polysome collapse and RNA condensation fluidize the cytoplasm. Authors: Ying Xie / Tong Shu / Tiewei Liu / Marie-Christin Spindler / Julia Mahamid / Glen M Hocky / David Gresham / Liam J Holt /   Abstract: The cell interior is packed with macromolecules of mesoscale size, and this crowded milieu significantly influences cellular physiology. Cellular stress responses almost universally lead to ...The cell interior is packed with macromolecules of mesoscale size, and this crowded milieu significantly influences cellular physiology. Cellular stress responses almost universally lead to inhibition of translation, resulting in polysome collapse and release of mRNA. The released mRNA molecules condense with RNA-binding proteins to form ribonucleoprotein (RNP) condensates known as processing bodies and stress granules. Here, we show that polysome collapse and condensation of RNA transiently fluidize the cytoplasm, and coarse-grained molecular dynamic simulations support this as a minimal mechanism for the observed biophysical changes. Increased mesoscale diffusivity correlates with the efficient formation of quality control bodies (Q-bodies), membraneless organelles that compartmentalize misfolded peptides during stress. Synthetic, light-induced RNA condensation also fluidizes the cytoplasm. Together, our study reveals a functional role for stress-induced translation inhibition and formation of RNP condensates in modulating the physical properties of the cytoplasm to enable efficient response of cells to stress conditions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50415.map.gz emd_50415.map.gz | 19.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50415-v30.xml emd-50415-v30.xml emd-50415.xml emd-50415.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50415_fsc.xml emd_50415_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_50415.png emd_50415.png | 60.6 KB | ||
| Masks |  emd_50415_msk_1.map emd_50415_msk_1.map | 20.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50415.cif.gz emd-50415.cif.gz | 4.1 KB | ||
| Others |  emd_50415_half_map_1.map.gz emd_50415_half_map_1.map.gz emd_50415_half_map_2.map.gz emd_50415_half_map_2.map.gz | 10.6 MB 10.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50415 http://ftp.pdbj.org/pub/emdb/structures/EMD-50415 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50415 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50415 | HTTPS FTP |
-Validation report
| Summary document |  emd_50415_validation.pdf.gz emd_50415_validation.pdf.gz | 856.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50415_full_validation.pdf.gz emd_50415_full_validation.pdf.gz | 856.4 KB | Display | |
| Data in XML |  emd_50415_validation.xml.gz emd_50415_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_50415_validation.cif.gz emd_50415_validation.cif.gz | 16.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50415 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50415 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50415 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50415 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50415.map.gz / Format: CCP4 / Size: 20.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50415.map.gz / Format: CCP4 / Size: 20.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of 80S ribosomes in S. cerevisiae under acute glucose starvation | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||
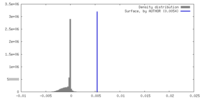
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50415_msk_1.map emd_50415_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
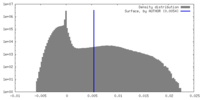
| Density Histograms |
-Half map: First half map of subtomogram average of 80S...
| File | emd_50415_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map of subtomogram average of 80S ribosomes in S. cerevisiae under acute glucose starvation | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-focused ion beam milled lamellae of S.cerevisiae cells under...
| Entire | Name: Cryo-focused ion beam milled lamellae of S.cerevisiae cells under acute glucose starvation |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-focused ion beam milled lamellae of S.cerevisiae cells under...
| Supramolecule | Name: Cryo-focused ion beam milled lamellae of S.cerevisiae cells under acute glucose starvation type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 2.11792 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 26000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)