+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pseudomonas aeruginosa MexY | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pseudomonsa aeruginosa / MexY / RND / Efflux / MEMBRANE PROTEIN | |||||||||
| Function / homology | Hydrophobe/amphiphile efflux-1 HAE1 / Acriflavin resistance protein / Multidrug efflux transporter AcrB TolC docking domain, DN/DC subdomains / AcrB/AcrD/AcrF family / efflux transmembrane transporter activity / xenobiotic transmembrane transporter activity / plasma membrane / Efflux pump membrane transporter Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
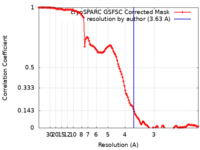
| Method | single particle reconstruction / cryo EM / Resolution: 3.63 Å | |||||||||
 Authors Authors | Gregor WD / Maharjan R / Zhang Z / Yu EW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Pseudomonas aeruginosa MexY Authors: Gregor WD / Maharjan R / Zhang Z / Yu EW | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47796.map.gz emd_47796.map.gz | 89.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47796-v30.xml emd-47796-v30.xml emd-47796.xml emd-47796.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_47796_fsc.xml emd_47796_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_47796.png emd_47796.png | 49.3 KB | ||
| Filedesc metadata |  emd-47796.cif.gz emd-47796.cif.gz | 5.9 KB | ||
| Others |  emd_47796_half_map_1.map.gz emd_47796_half_map_1.map.gz emd_47796_half_map_2.map.gz emd_47796_half_map_2.map.gz | 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47796 http://ftp.pdbj.org/pub/emdb/structures/EMD-47796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47796 | HTTPS FTP |
-Validation report
| Summary document |  emd_47796_validation.pdf.gz emd_47796_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_47796_full_validation.pdf.gz emd_47796_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_47796_validation.xml.gz emd_47796_validation.xml.gz | 20.9 KB | Display | |
| Data in CIF |  emd_47796_validation.cif.gz emd_47796_validation.cif.gz | 26.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47796 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47796 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47796 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47796 | HTTPS FTP |
-Related structure data
| Related structure data |  9e9fMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_47796.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47796.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_47796_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_47796_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MexY
| Entire | Name: MexY |
|---|---|
| Components |
|
-Supramolecule #1: MexY
| Supramolecule | Name: MexY / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Efflux pump membrane transporter
| Macromolecule | Name: Efflux pump membrane transporter / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 111.287914 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARFFIDRPV FAWVISLLIV LAGVLAIRFL PVAQYPDIAP PVVNVSASYP GASAKVVEEA VTAIIEREMN GAPGLLYTKA TSSTGQASL TLTFRQGVNA DLAAVEVQNR LKIVESRLPE SVRRDGIYVE KAADSIQLIV TLTSSSGRYD AMELGEIASS N VLQALRRV ...String: MARFFIDRPV FAWVISLLIV LAGVLAIRFL PVAQYPDIAP PVVNVSASYP GASAKVVEEA VTAIIEREMN GAPGLLYTKA TSSTGQASL TLTFRQGVNA DLAAVEVQNR LKIVESRLPE SVRRDGIYVE KAADSIQLIV TLTSSSGRYD AMELGEIASS N VLQALRRV EGVGKVETWG AEYAMRIWPD PAKLTSMNLS ASDLVNAVRR HNARLTVGDI GNLGVPDSAP ISATVKVDDT LV TPEQFGE IPLRIRADGG AIRLRDVARV EFGQSEYGFV SRVNQMTATG LAVKMAPGSN AVATAKRIRA TLDELSRYFP EGV SYNIPY DTSAFVEISI RKVVSTLLEA MLLVFAVMYL FMQNFRATLI PTLVVPVALL GTFTVMLGLG FSINVLTMFG MVLA IGILV DDAIIVVENV ERLMAEEGLS PHDATVKAMR QISGAIVGIT VVLVSVFVPM AFFSGAVGNI YRQFAVTLAV SIGFS AFLA LSLTPALCAT LLRPIDADHH EKRGFFGWFN RAFLRLTGRY RNAVAGILAR PIRWMLVYAL VIGVVALLFV RLPQAF LPE EDQGDFMIMV MQPEGTPMAE TMANVGDVER YLAEHEPVAY AYAVGGFSLY GDGTSSAMIF ATLKDWSERR EASQHVG AI VERINQRFAG LPNRTVYAMN SPPLPDLGST SGFDFRLQDR GGVGYEALVK ARDQLLARAA EDPRLANVMF AGQGEAPQ I RLDIDRRKAE TLGVSMDEIN TTLAVMFGSD YIGDFMHGSQ VRKVVVQADG AKRLGIDDIG RLHVRNEQGE MVPLATFAK AAWTLGPPQL TRYNGYPSFN LEGQAAPGYS SGEAMQAMEQ LMQGLPEGIA HEWSGQSFEE RLSGAQAPAL FALSVLIVFL ALAALYESW SIPLAVILVV PLGVLGALLG VSLRGLPNDI YFKVGLITII GLSAKNAILI IEVAKDHYQE GMSLLQATLE A ARLRLRPI VMTSLAFGFG VVPLALSSGA GSGAQVAIGT GVLGGIVTAT VLAVFLVPLF FLVVGRLFRL RKA UniProtKB: Efflux pump membrane transporter |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 37.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)