+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outward-facing Atorvastatin-bound OATP1B1 with sybody Sb5 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | solute carrier / sybody / membrane protein / drug-drug interactions / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLCO1B1 causes hyperbilirubinemia, Rotor type (HBLRR) / Transport of organic anions / sodium-independent organic anion transport / sodium-independent organic anion transmembrane transporter activity / thyroid hormone transmembrane transporter activity / prostaglandin transmembrane transporter activity / organic anion transport / heme catabolic process / organic anion transmembrane transporter activity / Atorvastatin ADME ...Defective SLCO1B1 causes hyperbilirubinemia, Rotor type (HBLRR) / Transport of organic anions / sodium-independent organic anion transport / sodium-independent organic anion transmembrane transporter activity / thyroid hormone transmembrane transporter activity / prostaglandin transmembrane transporter activity / organic anion transport / heme catabolic process / organic anion transmembrane transporter activity / Atorvastatin ADME / bile acid transmembrane transporter activity / Heme degradation / bile acid and bile salt transport / Recycling of bile acids and salts / monoatomic ion transport / xenobiotic metabolic process / basal plasma membrane / basolateral plasma membrane / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Sung MW / Lees JA / Han S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2025 Journal: J Biol Chem / Year: 2025Title: Cyclosporine A sterically inhibits statin transport by solute carrier OATP1B1. Authors: Min Woo Sung / Kuan Hu / Lea M Hurlimann / Joshua A Lees / Kimberly F Fennell / Mark A West / Chester Costales / Amilcar David Rodrigues / Iwan Zimmermann / Roger J P Dawson / Shenping Liu / Seungil Han /   Abstract: Members of the Organic Anion Transporter Polypeptides (OATP) are integral membrane proteins responsible for facilitating the transport of organic anions across the cell membrane. OATP1B1 (SLCO1B1), ...Members of the Organic Anion Transporter Polypeptides (OATP) are integral membrane proteins responsible for facilitating the transport of organic anions across the cell membrane. OATP1B1 (SLCO1B1), the prototypic OATP family member, is the most abundant uptake transporter in the liver and a key mediator of the hepatic uptake and clearance of numerous endogenous and xenobiotic compounds. It serves as a locus of important drug-drug interactions, such as those between statins and cyclosporine A, and carries the potential to enable liver-targeting therapeutics. In this study, we report cryo-EM structures of OATP1B1 and its complexes with one of its statin substrates, atorvastatin, and an inhibitor, cyclosporine A. This structural analysis has yielded insights into the mechanisms underlying the OATP1B1-mediated transport of statins and the inhibitory effect of cyclosporine A. These findings contribute to a better understanding of the molecular processes involved in drug transport and offer potential avenues for the development of targeted medications for liver-related conditions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_46005.map.gz emd_46005.map.gz | 49.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-46005-v30.xml emd-46005-v30.xml emd-46005.xml emd-46005.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_46005.png emd_46005.png | 65.8 KB | ||
| Filedesc metadata |  emd-46005.cif.gz emd-46005.cif.gz | 6.3 KB | ||
| Others |  emd_46005_half_map_1.map.gz emd_46005_half_map_1.map.gz emd_46005_half_map_2.map.gz emd_46005_half_map_2.map.gz | 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-46005 http://ftp.pdbj.org/pub/emdb/structures/EMD-46005 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-46005 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-46005 | HTTPS FTP |
-Validation report
| Summary document |  emd_46005_validation.pdf.gz emd_46005_validation.pdf.gz | 930.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_46005_full_validation.pdf.gz emd_46005_full_validation.pdf.gz | 930.2 KB | Display | |
| Data in XML |  emd_46005_validation.xml.gz emd_46005_validation.xml.gz | 11.7 KB | Display | |
| Data in CIF |  emd_46005_validation.cif.gz emd_46005_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-46005 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-46005 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-46005 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-46005 | HTTPS FTP |
-Related structure data
| Related structure data |  9cy3MC  9cy1C  9cy4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_46005.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_46005.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.099 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_46005_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_46005_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Outward-facing atorvastatin-bound OATP1B1 with Sybody 5
| Entire | Name: Outward-facing atorvastatin-bound OATP1B1 with Sybody 5 |
|---|---|
| Components |
|
-Supramolecule #1: Outward-facing atorvastatin-bound OATP1B1 with Sybody 5
| Supramolecule | Name: Outward-facing atorvastatin-bound OATP1B1 with Sybody 5 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier organic anion transporter family member 1B1
| Macromolecule | Name: Solute carrier organic anion transporter family member 1B1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 79.880977 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHHHHHHHH HDYKDDDDKE NLYFQGMDQN QHLNKTAEAQ PSENKKTRYC NGLKMFLAAL SLSFIAKTLG AIIMKSSIIH IERRFEISS SLVGFIDGSF EIGNLLVIVF VSYFGSKLHR PKLIGIGCFI MGIGGVLTAL PHFFMGYYRY SKETNINSSE N STSTLSTC ...String: MHHHHHHHHH HDYKDDDDKE NLYFQGMDQN QHLNKTAEAQ PSENKKTRYC NGLKMFLAAL SLSFIAKTLG AIIMKSSIIH IERRFEISS SLVGFIDGSF EIGNLLVIVF VSYFGSKLHR PKLIGIGCFI MGIGGVLTAL PHFFMGYYRY SKETNINSSE N STSTLSTC LINQILSLNR ASPEIVGKGC LKESGSYMWI YVFMGNMLRG IGETPIVPLG LSYIDDFAKE GHSSLYLGIL NA IAMIGPI IGFTLGSLFS KMYVDIGYVD LSTIRITPTD SRWVGAWWLN FLVSGLFSII SSIPFFFLPQ TPNKPQKERK ASL SLHVLE TNDEKDQTAN LTNQGKNITK NVTGFFQSFK SILTNPLYVM FVLLTLLQVS SYIGAFTYVF KYVEQQYGQP SSKA NILLG VITIPIFASG MFLGGYIIKK FKLNTVGIAK FSCFTAVMSL SFYLLYFFIL CENKSVAGLT MTYDGNNPVT SHRDV PLSY CNSDCNCDES QWEPVCGNNG ITYISPCLAG CKSSSGNKKP IVFYNCSCLE VTGLQNRNYS AHLGECPRDD ACTRKF YFF VAIQVLNLFF SALGGTSHVM LIVKIVQPEL KSLALGFHSM VIRALGGILA PIYFGALIDT TCIKWSTNNC GTRGSCR TY NSTSFSRVYL GLSSMLRVSS LVLYIILIYA MKKKYQEKDI NASENGSVMD EANLESLNKN KHFVPSAGAD SETHC UniProtKB: Solute carrier organic anion transporter family member 1B1 |
-Macromolecule #2: Sybody 5
| Macromolecule | Name: Sybody 5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.88551 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSSSQVQLVE SGGGLVQAGG SLRLSCAASG FPVNLSYMHW YRQAPGKERE WVAAISSWGW HTEYADSVKG RFTISRDNAK NTVYLQMNS LKPEDTAVYY CHVRVGRSYF GQGTQVSVSA GRAGEQKLIS EEDLNSAVDH HHHHH |
-Macromolecule #3: 7-[2-(4-FLUORO-PHENYL)-5-ISOPROPYL-3-PHENYL-4-PHENYLCARBAMOYL-PYR...
| Macromolecule | Name: 7-[2-(4-FLUORO-PHENYL)-5-ISOPROPYL-3-PHENYL-4-PHENYLCARBAMOYL-PYRROL-1-YL]- 3,5-DIHYDROXY-HEPTANOIC ACID type: ligand / ID: 3 / Number of copies: 1 / Formula: 117 |
|---|---|
| Molecular weight | Theoretical: 558.64 Da |
| Chemical component information | 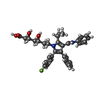 ChemComp-117: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































