[English] 日本語
 Yorodumi
Yorodumi- EMDB-45749: Cryo-EM structure of human claudin-4 complex with Clostridium per... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human claudin-4 complex with Clostridium perfringens enterotoxin, sFab COP-1, and Nanobody | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | claudin / Fab / enterotoxin / nanobody / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationparacellular transport / positive regulation of metallopeptidase activity / calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules / Tight junction interactions / bicellular tight junction assembly / apicolateral plasma membrane / regulation of cell morphogenesis / tight junction / positive regulation of wound healing / chloride channel activity ...paracellular transport / positive regulation of metallopeptidase activity / calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules / Tight junction interactions / bicellular tight junction assembly / apicolateral plasma membrane / regulation of cell morphogenesis / tight junction / positive regulation of wound healing / chloride channel activity / renal absorption / chloride channel complex / lateral plasma membrane / bicellular tight junction / establishment of skin barrier / basal plasma membrane / response to progesterone / female pregnancy / circadian rhythm / transmembrane signaling receptor activity / cell-cell junction / toxin activity / cell adhesion / positive regulation of cell migration / apical plasma membrane / structural molecule activity / extracellular region / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species | synthetic construct (others) /  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
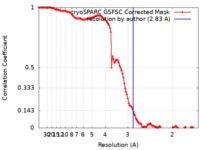
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | |||||||||
 Authors Authors | Vecchio AJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Cryo-EM structures of Clostridium perfringens enterotoxin bound to its human receptor, claudin-4. Authors: Sewwandi S Rathnayake / Satchal K Erramilli / Anthony A Kossiakoff / Alex J Vecchio /  Abstract: Clostridium perfringens enterotoxin (CpE) causes prevalent and deadly gastrointestinal disorders. CpE binds to receptors called claudins on the apical surfaces of small intestinal epithelium. ...Clostridium perfringens enterotoxin (CpE) causes prevalent and deadly gastrointestinal disorders. CpE binds to receptors called claudins on the apical surfaces of small intestinal epithelium. Claudins normally regulate paracellular transport but are hijacked from doing so by CpE and are instead led to form claudin/CpE complexes. Claudin/CpE complexes are the building blocks of oligomeric β-barrel pores that penetrate the plasma membrane and induce gut cytotoxicity. Here, we present the structures of CpE in complex with its native claudin receptor in humans, claudin-4, using cryogenic electron microscopy. The structures reveal the architecture of the claudin/CpE complex, the residues used in binding, the orientation of CpE relative to the membrane, and CpE-induced changes to claudin-4. Further, structures and modeling allude to the biophysical procession from claudin/CpE complexes to cytotoxic β-barrel pores during pathogenesis. In full, this work proposes a model of claudin/CpE assembly and provides strategies to obstruct its formation to treat CpE diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45749.map.gz emd_45749.map.gz | 88.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45749-v30.xml emd-45749-v30.xml emd-45749.xml emd-45749.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45749_fsc.xml emd_45749_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_45749.png emd_45749.png | 81.6 KB | ||
| Filedesc metadata |  emd-45749.cif.gz emd-45749.cif.gz | 7.3 KB | ||
| Others |  emd_45749_half_map_1.map.gz emd_45749_half_map_1.map.gz emd_45749_half_map_2.map.gz emd_45749_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45749 http://ftp.pdbj.org/pub/emdb/structures/EMD-45749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45749 | HTTPS FTP |
-Validation report
| Summary document |  emd_45749_validation.pdf.gz emd_45749_validation.pdf.gz | 767.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_45749_full_validation.pdf.gz emd_45749_full_validation.pdf.gz | 767 KB | Display | |
| Data in XML |  emd_45749_validation.xml.gz emd_45749_validation.xml.gz | 20.6 KB | Display | |
| Data in CIF |  emd_45749_validation.cif.gz emd_45749_validation.cif.gz | 26.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45749 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45749 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45749 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45749 | HTTPS FTP |
-Related structure data
| Related structure data |  9cmiMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45749.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45749.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_45749_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_45749_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human claudin-4 complex with Clostridium perfringens enterotoxin,...
| Entire | Name: Human claudin-4 complex with Clostridium perfringens enterotoxin, sFab COP-1, and Nanobody |
|---|---|
| Components |
|
-Supramolecule #1: Human claudin-4 complex with Clostridium perfringens enterotoxin,...
| Supramolecule | Name: Human claudin-4 complex with Clostridium perfringens enterotoxin, sFab COP-1, and Nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 / Details: 5-protein complex |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: Claudin-4
| Macromolecule | Name: Claudin-4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.234332 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSMASMGLQV MGIALAVLGW LAVMLCCALP MWRVTAFIGS NIVTSQTIWE GLWMNCVVQS TGQMQCKVYD SLLALPQDLQ AARALVIIS IIVAALGVLL SVVGGKCTNC LEDESAKAKT MIVAGVVFLL AGLMVIVPVS WTAHNIIQDF YNPLVASGQK R EMGASLYV ...String: GSMASMGLQV MGIALAVLGW LAVMLCCALP MWRVTAFIGS NIVTSQTIWE GLWMNCVVQS TGQMQCKVYD SLLALPQDLQ AARALVIIS IIVAALGVLL SVVGGKCTNC LEDESAKAKT MIVAGVVFLL AGLMVIVPVS WTAHNIIQDF YNPLVASGQK R EMGASLYV GWAASGLLLL GGGLLCCNCP PRTDKPYSAK YSAARSAAAS NYV UniProtKB: Claudin-4 |
-Macromolecule #2: Heat-labile enterotoxin B chain
| Macromolecule | Name: Heat-labile enterotoxin B chain / type: protein_or_peptide / ID: 2 / Details: Trypsin treated CpE / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.000582 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: TPINITNSNS NLSDGLYVID KGDGWILGEP SVVSSQILNP NETGTFSQSL TKSKEVSINV NFSVGFTSEF IQASVEYGFG ITIGEQNTI ERSVSTTAGP NEYVYYKVYA TYRKYQAIRI SHGNISDDGS IYKLTGIWLS KTSADSLGNI DQGSLIETGE R CVLTVPST ...String: TPINITNSNS NLSDGLYVID KGDGWILGEP SVVSSQILNP NETGTFSQSL TKSKEVSINV NFSVGFTSEF IQASVEYGFG ITIGEQNTI ERSVSTTAGP NEYVYYKVYA TYRKYQAIRI SHGNISDDGS IYKLTGIWLS KTSADSLGNI DQGSLIETGE R CVLTVPST DIEKEILDLA AATERLNLTD ALNSNPAGNL YDWRSSNSYP WTQKLNLHLT ITATGQKYRI LASKIVDFNI YS NNFNNLV KLEQSLGDGV KDHYVDISLD AGQYVLVMKA NSSYSGNYPY SILFQKFGLV PR UniProtKB: Heat-labile enterotoxin B chain |
-Macromolecule #3: COP-1 sFab Heavy Chain
| Macromolecule | Name: COP-1 sFab Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.064498 KDa |
| Sequence | String: MKKNIAFLLA SMFVFSIATN AYAEISEVQL VESGGGLVQP GGSLRLSCAA SGFNFSSSYI HWVRQAPGKG LEWVASISSS SGSTSYADS VKGRFTISAD TSKNTAYLQM NSLRAEDTAV YYCARWFHPW WWWEYLFRGA IDYWGQGTLV TVSSASTKGP S VFPLAPSS ...String: MKKNIAFLLA SMFVFSIATN AYAEISEVQL VESGGGLVQP GGSLRLSCAA SGFNFSSSYI HWVRQAPGKG LEWVASISSS SGSTSYADS VKGRFTISAD TSKNTAYLQM NSLRAEDTAV YYCARWFHPW WWWEYLFRGA IDYWGQGTLV TVSSASTKGP S VFPLAPSS KSTSGGTAAL GCLVKDYFPE PVTVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NH KPSNTKV DKKVEPKSCD KTHT |
-Macromolecule #4: Anti-Fab Nanobody
| Macromolecule | Name: Anti-Fab Nanobody / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.175438 KDa |
| Sequence | String: GSVQLQESGG GLVQPGGSLR LSCAASGRTI SRYAMSWFRQ APGKEREFVA VARRSGDGAF YADSVQGRFT VSRDDAKNTV YLQMNSLKP EDTAVYYCAI DSDTFYSGSY DYWGQGTQVT VS |
-Macromolecule #5: COP-1 sFab Light Chain
| Macromolecule | Name: COP-1 sFab Light Chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.794859 KDa |
| Sequence | String: MKKNIAFLLA SMFVFSIATN AYASDIQMTQ SPSSLSASVG DRVTITCRAS QSVSSAVAWY QQKPGKAPKL LIYSASSLYS GVPSRFSGS RSGTDFTLTI SSLQPEDFAT YYCQQSSSSL ITFGQGTKVE IKRTVAAPSV FIFPPSDSQL KSGTASVVCL L NNFYPREA ...String: MKKNIAFLLA SMFVFSIATN AYASDIQMTQ SPSSLSASVG DRVTITCRAS QSVSSAVAWY QQKPGKAPKL LIYSASSLYS GVPSRFSGS RSGTDFTLTI SSLQPEDFAT YYCQQSSSSL ITFGQGTKVE IKRTVAAPSV FIFPPSDSQL KSGTASVVCL L NNFYPREA KVQWKVDNAL QSGNSQESVT EQDSKDSTYS LSSTLTLSKA DYEKHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Macromolecule #6: Lauryl Maltose Neopentyl Glycol
| Macromolecule | Name: Lauryl Maltose Neopentyl Glycol / type: ligand / ID: 6 / Number of copies: 1 / Formula: AV0 |
|---|---|
| Molecular weight | Theoretical: 1.005188 KDa |
| Chemical component information |  ChemComp-AV0: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.6 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 20 mM Hepes pH 7.4, 100 mM NaCl, and 0.003% LMNG | ||||||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. Details: glow-discharged for 30 seconds at 20 W using a Solarus 950 (Gatan) plasma cleaner | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Software | Name: EPU |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6774 / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Software | Name:  UCSF Chimera UCSF Chimera | ||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-9cmi: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)