[English] 日本語
 Yorodumi
Yorodumi- EMDB-45580: Cryo-EM structure of the Nipah Virus polymerase (L) protein in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Nipah Virus polymerase (L) protein in complex with the tetrameric phosphoprotein (P) | |||||||||
 Map data Map data | The NiV L-P map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nipah virus / L protein / phosphoprotein / RNA-dependent RNA polymerase / PRNTase / GDP polyribonucleotidyl transferase / RNA capping / viral replication / TRANSFERASE / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative stranded viral RNA transcription / NNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / virion component / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm / molecular adaptor activity / symbiont-mediated suppression of host innate immune response ...negative stranded viral RNA transcription / NNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / virion component / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm / molecular adaptor activity / symbiont-mediated suppression of host innate immune response / RNA-directed RNA polymerase / RNA-dependent RNA polymerase activity / GTPase activity / virus-mediated perturbation of host defense response / ATP binding Similarity search - Function | |||||||||
| Biological species |  Henipavirus nipahense Henipavirus nipahense | |||||||||
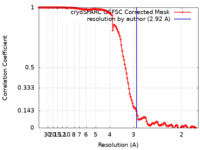
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Liu B / Yang G / Wang D | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure of the Nipah virus polymerase phosphoprotein complex. Authors: Ge Yang / Dong Wang / Bin Liu /  Abstract: The Nipah virus (NiV), a member of the Paramyxoviridae family, is notorious for its high fatality rate in humans. The RNA polymerase machinery of NiV, comprising the large protein L and the ...The Nipah virus (NiV), a member of the Paramyxoviridae family, is notorious for its high fatality rate in humans. The RNA polymerase machinery of NiV, comprising the large protein L and the phosphoprotein P, is essential for viral replication. This study presents the 2.9-Å cryo-electron microscopy structure of the NiV L-P complex, shedding light on its assembly and functionality. The structure not only demonstrates the molecular details of the conserved N-terminal domain, RNA-dependent RNA polymerase (RdRp), and GDP polyribonucleotidyltransferase of the L protein, but also the intact central oligomerization domain and the C-terminal X domain of the P protein. The P protein interacts extensively with the L protein, forming an antiparallel β-sheet among the P protomers and with the fingers subdomain of RdRp. The flexible linker domain of one P promoter extends its contact with the fingers subdomain to reach near the nascent RNA exit, highlighting the distinct characteristic of the NiV L-P interface. This distinctive tetrameric organization of the P protein and its interaction with the L protein provide crucial molecular insights into the replication and transcription mechanisms of NiV polymerase, ultimately contributing to the development of effective treatments and preventive measures against this Paramyxoviridae family deadly pathogen. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45580.map.gz emd_45580.map.gz | 108.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45580-v30.xml emd-45580-v30.xml emd-45580.xml emd-45580.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45580_fsc.xml emd_45580_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_45580.png emd_45580.png | 88 KB | ||
| Filedesc metadata |  emd-45580.cif.gz emd-45580.cif.gz | 7.9 KB | ||
| Others |  emd_45580_additional_1.map.gz emd_45580_additional_1.map.gz emd_45580_half_map_1.map.gz emd_45580_half_map_1.map.gz emd_45580_half_map_2.map.gz emd_45580_half_map_2.map.gz | 179.3 MB 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45580 http://ftp.pdbj.org/pub/emdb/structures/EMD-45580 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45580 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45580 | HTTPS FTP |
-Validation report
| Summary document |  emd_45580_validation.pdf.gz emd_45580_validation.pdf.gz | 878.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_45580_full_validation.pdf.gz emd_45580_full_validation.pdf.gz | 878.2 KB | Display | |
| Data in XML |  emd_45580_validation.xml.gz emd_45580_validation.xml.gz | 21.1 KB | Display | |
| Data in CIF |  emd_45580_validation.cif.gz emd_45580_validation.cif.gz | 26.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45580 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45580 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45580 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45580 | HTTPS FTP |
-Related structure data
| Related structure data |  9cgiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45580.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45580.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The NiV L-P map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88533 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : The Nipah virus L-P complex
| Entire | Name: The Nipah virus L-P complex |
|---|---|
| Components |
|
-Supramolecule #1: The Nipah virus L-P complex
| Supramolecule | Name: The Nipah virus L-P complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Henipavirus nipahense Henipavirus nipahense |
| Molecular weight | Theoretical: 650 KDa |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Henipavirus nipahense Henipavirus nipahense |
| Molecular weight | Theoretical: 257.565156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADELSISDI IYPECHLDSP IVSGKLISAI EYAQLRHNQP SDDKRLSENI RLNLHGKRKS LYILRQSKQG DYIRNNIKNL KEFMHIAYP ECNNILFSIT SQGMTSKLDN IMKKSFKAYN IISKKVIGML QNITRNLITQ DRRDEIINIH ECRRLGDLGK N MSQSKWYE ...String: MADELSISDI IYPECHLDSP IVSGKLISAI EYAQLRHNQP SDDKRLSENI RLNLHGKRKS LYILRQSKQG DYIRNNIKNL KEFMHIAYP ECNNILFSIT SQGMTSKLDN IMKKSFKAYN IISKKVIGML QNITRNLITQ DRRDEIINIH ECRRLGDLGK N MSQSKWYE CFLFWFTIKT EMRAVIKNSQ KPKFRSDSCI IHMRDKSTEI ILNPNLICIF KSDKTGKKCY YLTPEMVLMY CD VLEGRMM METTVKSDIK YQPLISRSNA LWGLIDPLFP VMGNRIYNIV SMIEPLVLAL LQLKDEARIL RGAFLHHCIK EMH QELSEC GFTDQKIRSM FIDDLLSILN IDNIHLLAEF FSFFRTFGHP ILEAKVAAEK VREHMLADKV LEYAPIMKAH AIFC GTIIN GYRDRHGGAW PPLYLPAHAS KHIIRLKNSG ESLTIDDCVK NWESFCGIQF DCFMELKLDS DLSMYMKDKA LSPIK DEWD SVYPREVLSY TPPKSTEPRR LVDVFVNDEN FDPYNMLEYV LSGAYLEDEQ FNVSYSLKEK ETKQAGRLFA KMTYKM RAC QVIAEALIAS GVGKYFKENG MVKDEHELLK TLFQLSISSV PRGNSQGNDP QSINNIERDF QYFKGVTTNV KDKKNNS FN KVKSALNNPC QADGVHHNMS PNTRNRYKCS NTSKSFLDYH TEFNPHNHYK SDNTEAAVLS RYEDNTGTKF DTVSAFLT T DLKKFCLNWR YESMAIFAER LDEIYGLPGF FNWMHKRLER SVIYVADPNC PPNIDKHMEL EKTPEDDIFI HYPKGGIEG YSQKTWTIAT IPFLFLSAYE TNTRIAAIVQ GDNESIAITQ KVHPNLPYKV KKEICAKQAQ LYFERLRMNL RALGHNLKAT ETIISTHLF IYSKKIHYDG AVLSQALKSM SRCCFWSETL VDETRSACSN ISTTIAKAIE NGLSRNVGYC INILKVIQQL L ISTEFSIN ETLTLDVTSP ISNNLDWLIT AALIPAPIGG FNYLNLSRIF VRNIGDPVTA SLADLKRMID HSIMTESVLQ KV MNQEPGD ASFLDWASDP YSGNLPDSQS ITKTIKNITA RTILRNSPNP MLKGLFHDKS FDEDLELASF LMDRRVILPR AAH EILDNS LTGAREEIAG LLDTTKGLIR SGLRKSGLQP KLVSRLSHHD YNQFLILNKL LSNRRQNDLI SSNTCSVDLA RALR SHMWR ELALGRVIYG LEVPDALEAM VGRYITGSLE CQICEQGNTM YGWFFVPRDS QLDQVDREHS SIRVPYVGSS TDERS DIKL GNVKRPTKAL RSAIRIATVY TWAYGDNEEC WYEAWYLASQ RVNIDLDVLK AITPVSTSNN LSHRLRDKST QFKFAG SVL NRVSRYVNIS NDNLDFRIEG EKVDTNLIYQ QAMLLGLSVL EGKFRLRLET DDYNGIYHLH VKDNCCVKEV ADVGQVD AE LPIPEYTEVD NNHLIYDPDP VSEIDCSRLS NQESKSRELD FPLWSTEELH DVLAKTVAQT VLEIITKADK DVLKQHLA I DSDDNINSLI TEFLIVDPEL FALYLGQSIS IKWAFEIHHR RPRGRHTMVD LLSDLVSNTS KHTYKVLSNA LSHPRVFKR FVNCGLLLPT QGPYLHQQDF EKLSQNLLVT SYMIYLMNWC DFKKSPFLIA EQDETVISLR EDIITSKHLC VIIDLYANHH KPPWIIDLN PQEKICVLRD FISKSRHVDT SSRSWNTSDL DFVIFYASLT YLRRGIIKQL RIRQVTEVID TTTMLRDNII V ENPPIKTG VLDIRGCIIY NLEEILSMNT KSASKKIFNL NSRPSVENHK YRRIGLNSSS CYKALNLSPL IQRYLPSGAQ RL FIGEGSG SMMLLYQSTL GQSISFYNSG IDGDYIPGQR ELKLFPSEYS IAEEDPSLTG KLKGLVVPLF NGRPETTWIG NLD SYEYII NRTAGRSIGL VHSDMESGID KNVEEILVEH SHLISIAINV MMEDGLLVSK IAYTPGFPIS RLFNMYRSYF GLVL VCFPV YSNPDSTEVY LLCLQKTVKT IVPPQKVLEH SNLHDEVNDQ GITSVIFKIK NSQSKQFHDD LKKYYQIDQP FFVPT KITS DEQVLLQAGL KLNGPEILKS EISYDIGSDI NTLRDTIIIM LNEAMNYFDD NRSPSHHLEP YPVLERTRIK TIMNCV TKK VIVYSLIKFK DTKSSELYHI KNNIRRKVLI LDFRSKLMTK TLPKGMQERR EKNGFKEVWI VDLSNREVKI WWKIIGY IS II UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: Phosphoprotein
| Macromolecule | Name: Phosphoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Henipavirus nipahense Henipavirus nipahense |
| Molecular weight | Theoretical: 78.39032 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDKLELVNDG LNIIDFIQKN QKEIQKTYGR SSIQQPSIKD QTKAWEDFLQ CTSGESEQVE GGMSKDDGDV ERRNLEDLSS TSPTDGTIG KRVSNTRDWA EGSDDIQLDP VVTDVVYHDH GGECTGYGFT SSPERGWSDY TSGANNGNVC LVSDAKMLSY A PEIAVSKE ...String: MDKLELVNDG LNIIDFIQKN QKEIQKTYGR SSIQQPSIKD QTKAWEDFLQ CTSGESEQVE GGMSKDDGDV ERRNLEDLSS TSPTDGTIG KRVSNTRDWA EGSDDIQLDP VVTDVVYHDH GGECTGYGFT SSPERGWSDY TSGANNGNVC LVSDAKMLSY A PEIAVSKE DRETDLVHLE NKLSTTGLNP TAVPFTLRNL SDPAKDSPVI AEHYYGLGVK EQNVGPQTSR NVNLDSIKLY TS DDEEADQ LEFEDEFAGS SSEVIVGISP EDEEPSSVGG KPNESIGRTI EGQSIRDNLQ AKDNKSTDVP GAGPKDSAVK EEP PQKRLP MLAEEFECSG SEDPIIRELL KENSLINCQQ GKDAQPPYHW SIERSISPDK TEIVNGAVQT ADRQRPGTPM PKSR GIPIK KGTDAKYPSA GTENVPGSKS GATRHVRGSP PYQEGKSVNA ENVQLNASTA VKETDKSEVN PVDDNDSLDD KYIMP SDDF SNTFFPHDTD RLNYHADHLG DYDLETLCEE SVLMGVINSI KLINLDMRLN HIEEQVKEIP KIINKLESID RVLAKT NTA LSTIEGHLVS MMIMIPGKGK GERKGKNNPE LKPVIGRDIL EQQSLFSFDN VKNFRDGSLT NEPYGAAVQL REDLILP EL NFEETNASQF VPMADDSSRD VIKTLIRTHI KDRELRSELI GYLNKAENDE EIQEIANTVN DIIDGNI UniProtKB: Phosphoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: 50 mM Tris-HCl pH 8.0, 250 mM NaCl, 5% glycerol, 1 mM TCEP, and 4 mM MgCl2 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 63.0 K / Max: 77.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 6114 / Average exposure time: 1.7 sec. / Average electron dose: 54.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 73.5 |
|---|---|
| Output model |  PDB-9cgi: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)