[English] 日本語
 Yorodumi
Yorodumi- EMDB-45425: Cryo-EM structure of epinephrine-bound alpha-2A-adrenergic recept... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of epinephrine-bound alpha-2A-adrenergic receptor in complex with heterotrimeric Gi-protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Adrenergic Receptor / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of uterine smooth muscle contraction / adenylate cyclase-inhibiting adrenergic receptor signaling pathway / phospholipase C-activating adrenergic receptor signaling pathway / alpha2-adrenergic receptor activity / Adrenaline signalling through Alpha-2 adrenergic receptor / alpha-2C adrenergic receptor binding / receptor transactivation / epinephrine binding / alpha-1B adrenergic receptor binding / negative regulation of norepinephrine secretion ...negative regulation of uterine smooth muscle contraction / adenylate cyclase-inhibiting adrenergic receptor signaling pathway / phospholipase C-activating adrenergic receptor signaling pathway / alpha2-adrenergic receptor activity / Adrenaline signalling through Alpha-2 adrenergic receptor / alpha-2C adrenergic receptor binding / receptor transactivation / epinephrine binding / alpha-1B adrenergic receptor binding / negative regulation of norepinephrine secretion / Extra-nuclear estrogen signaling / negative regulation of calcium ion transmembrane transporter activity / Adenylate cyclase inhibitory pathway / negative regulation of epinephrine secretion / Olfactory Signaling Pathway / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / heterotrimeric G-protein binding / negative regulation of calcium ion-dependent exocytosis / fear response / dopaminergic synapse / Surfactant metabolism / positive regulation of potassium ion transport / Activation of the phototransduction cascade / thermoception / thioesterase binding / negative regulation of insulin secretion involved in cellular response to glucose stimulus / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / GTPase activating protein binding / negative regulation of synaptic transmission / positive regulation of membrane protein ectodomain proteolysis / norepinephrine binding / Adrenoceptors / intestinal absorption / G alpha (i) signalling events / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Glucagon-type ligand receptors / G alpha (12/13) signalling events / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Thrombin signalling through proteinase activated receptors (PARs) / Ca2+ pathway / Extra-nuclear estrogen signaling / G alpha (z) signalling events / G alpha (s) signalling events / positive regulation of epidermal growth factor receptor signaling pathway / neurotransmitter receptor localization to postsynaptic specialization membrane / G alpha (q) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (i) signalling events / positive regulation of wound healing / activation of protein kinase activity / Vasopressin regulates renal water homeostasis via Aquaporins / adrenergic receptor signaling pathway / negative regulation of calcium ion transport / Rho protein signal transduction / regulation of vasoconstriction / D2 dopamine receptor binding / negative regulation of lipid catabolic process / positive regulation of protein localization to cell cortex / negative regulation of insulin secretion / regulation of cAMP-mediated signaling / G protein-coupled serotonin receptor binding / adenylate cyclase-activating adrenergic receptor signaling pathway / GABA-ergic synapse / cellular response to forskolin / presynaptic active zone membrane / viral release from host cell by cytolysis / axon terminus / cellular response to hormone stimulus / regulation of mitotic spindle organization / presynaptic modulation of chemical synaptic transmission / activation of protein kinase B activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / peptidoglycan catabolic process / guanyl-nucleotide exchange factor activity / positive regulation of cytokine production / female pregnancy / G protein-coupled receptor binding / postsynaptic density membrane / positive regulation of MAP kinase activity / G-protein beta/gamma-subunit complex binding / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / platelet activation / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Lou JS / Su M / Wang J / Do HN / Miao Y / Huang XY | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Exp Mol Med / Year: 2024 Journal: Exp Mol Med / Year: 2024Title: Distinct binding conformations of epinephrine with α- and β-adrenergic receptors. Authors: Jian-Shu Lou / Minfei Su / Jinan Wang / Hung Nguyen Do / Yinglong Miao / Xin-Yun Huang /   Abstract: Agonists targeting α-adrenergic receptors (ARs) are used to treat diverse conditions, including hypertension, attention-deficit/hyperactivity disorder, pain, panic disorders, opioid and alcohol ...Agonists targeting α-adrenergic receptors (ARs) are used to treat diverse conditions, including hypertension, attention-deficit/hyperactivity disorder, pain, panic disorders, opioid and alcohol withdrawal symptoms, and cigarette cravings. These receptors transduce signals through heterotrimeric Gi proteins. Here, we elucidated cryo-EM structures that depict α-AR in complex with Gi proteins, along with the endogenous agonist epinephrine or the synthetic agonist dexmedetomidine. Molecular dynamics simulations and functional studies reinforce the results of the structural revelations. Our investigation revealed that epinephrine exhibits different conformations when engaging with α-ARs and β-ARs. Furthermore, α-AR and β-AR (primarily coupled to Gs, with secondary associations to Gi) were compared and found to exhibit different interactions with Gi proteins. Notably, the stability of the epinephrine-α-AR-Gi complex is greater than that of the dexmedetomidine-α-AR-Gi complex. These findings substantiate and improve our knowledge on the intricate signaling mechanisms orchestrated by ARs and concurrently shed light on the regulation of α-ARs and β-ARs by epinephrine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45425.map.gz emd_45425.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45425-v30.xml emd-45425-v30.xml emd-45425.xml emd-45425.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_45425.png emd_45425.png | 76 KB | ||
| Filedesc metadata |  emd-45425.cif.gz emd-45425.cif.gz | 6.4 KB | ||
| Others |  emd_45425_half_map_1.map.gz emd_45425_half_map_1.map.gz emd_45425_half_map_2.map.gz emd_45425_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45425 http://ftp.pdbj.org/pub/emdb/structures/EMD-45425 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45425 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45425 | HTTPS FTP |
-Validation report
| Summary document |  emd_45425_validation.pdf.gz emd_45425_validation.pdf.gz | 756.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_45425_full_validation.pdf.gz emd_45425_full_validation.pdf.gz | 756.5 KB | Display | |
| Data in XML |  emd_45425_validation.xml.gz emd_45425_validation.xml.gz | 13.1 KB | Display | |
| Data in CIF |  emd_45425_validation.cif.gz emd_45425_validation.cif.gz | 15.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45425 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45425 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45425 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45425 | HTTPS FTP |
-Related structure data
| Related structure data |  9cblMC  9cbmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45425.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45425.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
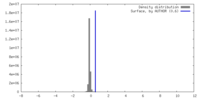
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Epinephrine-bound alpha-2A-adrenergic receptor in complex with he...
| Entire | Name: Epinephrine-bound alpha-2A-adrenergic receptor in complex with heterotrimeric Gi-protein |
|---|---|
| Components |
|
-Supramolecule #1: Epinephrine-bound alpha-2A-adrenergic receptor in complex with he...
| Supramolecule | Name: Epinephrine-bound alpha-2A-adrenergic receptor in complex with heterotrimeric Gi-protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.14707 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLEVLFQG PHMASMGCTL SAEDKAAVER SKMIDRNLRE DGEKAAREVK LLLLGAGESG KSTIVKQMKI IHEAGYSEE ECKQYKAVVY SNTIQSIIAI IRAMGRLKID FGDAARADDA RQLFVLAGAA EEGFMTAELA GVIKRLWKDS G VQACFNRS ...String: MGSSHHHHHH SSGLEVLFQG PHMASMGCTL SAEDKAAVER SKMIDRNLRE DGEKAAREVK LLLLGAGESG KSTIVKQMKI IHEAGYSEE ECKQYKAVVY SNTIQSIIAI IRAMGRLKID FGDAARADDA RQLFVLAGAA EEGFMTAELA GVIKRLWKDS G VQACFNRS REYQLNDSAA YYLNDLDRIA QPNYIPTQQD VLRTRVKTTG IVETHFTFKD LHFKMFDVGA QRSERKKWIH CF EGVTAII FCVALSDYDL VLAEDEEMNR MHESMKLFDS ICNNKWFTDT SIILFLNKKD LFEEKIKKSP LTICYPEYAG SNT YEEAAA YIQCQFEDLN KRKDTKEIYT HFTCATDTKN VQFVFDAVTD VIIKNNLKDC GLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.285734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW ...String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NAICFFPNGN AF ATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGHDNRVSCL GVT DDGMAV ATGSWDSFLK IWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.845078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFSAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Endolysin,Alpha-2A adrenergic receptor
| Macromolecule | Name: Endolysin,Alpha-2A adrenergic receptor / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: lysozyme |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.715871 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKNIFEML RIDEGLRLKI YKDTEGYYTI GIGHLLTKSP SLNAAKSELD KAIGRNTNGV ITKDEAEKL FNQDVDAAVR GILRNAKLKP VYDSLDAVRR AALINMVFQM GETGVAGFTN SLRMLQQKRW DEAAVNLAKS R WYNQTPNR ...String: MKTIIALSYI FCLVFADYKD DDDKNIFEML RIDEGLRLKI YKDTEGYYTI GIGHLLTKSP SLNAAKSELD KAIGRNTNGV ITKDEAEKL FNQDVDAAVR GILRNAKLKP VYDSLDAVRR AALINMVFQM GETGVAGFTN SLRMLQQKRW DEAAVNLAKS R WYNQTPNR AKRVITTFRT GTWDAYAAAG GGARATPYSL QVTLTLVCLA GLLMLLTVFG NVLVIIAVFT SRALKAPQNL FL VSLASAD ILVATLVIPF SLANEVMGYW YFGKAWCEIY LALDVLFCTS SIVHLCAISL DRYWSITQAI EYNLKRTPRR IKA IIITVW VISAVISFPP LISIEKKGGG GGPQPAEPRC EINDQKWYVI SSCIGSFFAP CLIMILVYVR IYQIAKRRTR QNRE KRFTF VLAVVIGVFV VCWFPFFFTY TLTAVGCSVP RTLFKFFFWF GYCNSSLNPV IYTIFNHDFR RAFKKILCRG DASLE VLFQ UniProtKB: Endolysin, Alpha-2A adrenergic receptor, Alpha-2A adrenergic receptor |
-Macromolecule #5: L-EPINEPHRINE
| Macromolecule | Name: L-EPINEPHRINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: ALE |
|---|---|
| Molecular weight | Theoretical: 183.204 Da |
| Chemical component information |  ChemComp-ALE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 713558 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)