+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of Azo-ffsy fiber | |||||||||
 Map data Map data | cryo-EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | D-peptide / peptide-fiber / helical / PROTEIN FIBRIL | |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Zia A / Guo J / Xu B / Wang F | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Am Chem Soc / Year: 2024 Journal: J Am Chem Soc / Year: 2024Title: Cell-Free Nonequilibrium Assembly for Hierarchical Protein/Peptide Nanopillars. Authors: Jiaqi Guo / Ayisha Zia / Qianfeng Qiu / Michael Norton / Kangqiang Qiu / Junichi Usuba / Zhiyu Liu / Meihui Yi / Shane T Rich-New / Michael Hagan / Seth Fraden / Grace D Han / Jiajie Diao / ...Authors: Jiaqi Guo / Ayisha Zia / Qianfeng Qiu / Michael Norton / Kangqiang Qiu / Junichi Usuba / Zhiyu Liu / Meihui Yi / Shane T Rich-New / Michael Hagan / Seth Fraden / Grace D Han / Jiajie Diao / Fengbin Wang / Bing Xu /  Abstract: Cells contain intricate protein nanostructures, but replicating them outside of cells presents challenges. One such example is the vertical fibronectin pillars observed in embryos. Here, we ...Cells contain intricate protein nanostructures, but replicating them outside of cells presents challenges. One such example is the vertical fibronectin pillars observed in embryos. Here, we demonstrate the creation of cell-free vertical fibronectin pillar mimics using nonequilibrium self-assembly. Our approach utilizes enzyme-responsive phosphopeptides that assemble into nanotubes. Enzyme action triggers shape changes in peptide assemblies, driving the vertical growth of protein nanopillars into bundles. These bundles, with peptide nanotubes serving as a template to remodel fibronectin, can then recruit collagen, which forms aggregates or bundles depending on their types. Nanopillar formation relies on enzyme-catalyzed nonequilibrium self-assembly and is governed by the concentrations of enzyme, protein, peptide, the structure of the peptide, and peptide assembly morphologies. Cryo-EM reveals unexpected nanotube thinning and packing after dephosphorylation, indicating a complex sculpting process during assembly. Our study demonstrates a cell-free method for constructing intricate, multiprotein nanostructures with directionality and composition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44637.map.gz emd_44637.map.gz | 4.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44637-v30.xml emd-44637-v30.xml emd-44637.xml emd-44637.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44637.png emd_44637.png | 52.4 KB | ||
| Filedesc metadata |  emd-44637.cif.gz emd-44637.cif.gz | 4.4 KB | ||
| Others |  emd_44637_half_map_1.map.gz emd_44637_half_map_1.map.gz emd_44637_half_map_2.map.gz emd_44637_half_map_2.map.gz | 115 MB 115 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44637 http://ftp.pdbj.org/pub/emdb/structures/EMD-44637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44637 | HTTPS FTP |
-Validation report
| Summary document |  emd_44637_validation.pdf.gz emd_44637_validation.pdf.gz | 682.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44637_full_validation.pdf.gz emd_44637_full_validation.pdf.gz | 682 KB | Display | |
| Data in XML |  emd_44637_validation.xml.gz emd_44637_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_44637_validation.cif.gz emd_44637_validation.cif.gz | 16.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44637 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44637 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44637 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44637 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44637.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44637.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
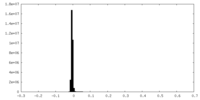
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: cryo-EM half map
| File | emd_44637_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
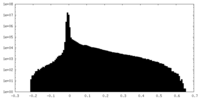
| Density Histograms |
-Half map: cryo-EM half map
| File | emd_44637_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
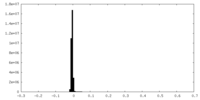
| Density Histograms |
- Sample components
Sample components
-Entire : ffsy fiber
| Entire | Name: ffsy fiber |
|---|---|
| Components |
|
-Supramolecule #1: ffsy fiber
| Supramolecule | Name: ffsy fiber / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: D-peptide ffsy
| Macromolecule | Name: D-peptide ffsy / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 770.83 Da |
| Sequence | String: (A1APX)(DPN)(DPN)(DSN)(DTY) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
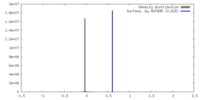
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 1.312 Å Applied symmetry - Helical parameters - Δ&Phi: 63.370 ° Applied symmetry - Helical parameters - Axial symmetry: C2 (2 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 3485262 |
|---|---|
| Startup model | Type of model: NONE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)