+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the ZBTB5 BTB domain filament | |||||||||
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | BTB domain / transcription factor / ZBTB protein / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase II transcription regulatory region sequence-specific DNA binding / DNA-binding transcription repressor activity, RNA polymerase II-specific / DNA-binding transcription factor activity, RNA polymerase II-specific / regulation of transcription by RNA polymerase II / chromatin / negative regulation of transcription by RNA polymerase II / metal ion binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
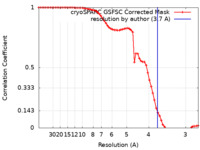
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Park J / Hunkeler M / Fischer ES | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Polymerization of ZBTB transcription factors regulates chromatin occupancy. Authors: Paul M C Park / Jiho Park / Jared Brown / Moritz Hunkeler / Shourya S Roy Burman / Katherine A Donovan / Hojong Yoon / Radosław P Nowak / Mikołaj Słabicki / Benjamin L Ebert / Eric S Fischer /  Abstract: BCL6, an oncogenic transcription factor (TF), forms polymers in the presence of a small-molecule molecular glue that stabilizes a complementary interface between homodimers of BCL6's broad-complex, ...BCL6, an oncogenic transcription factor (TF), forms polymers in the presence of a small-molecule molecular glue that stabilizes a complementary interface between homodimers of BCL6's broad-complex, tramtrack, and bric-à-brac (BTB) domain. The BTB domains of other proteins, including a large class of TFs, have similar architectures and symmetries, raising the possibility that additional BTB proteins self-assemble into higher-order structures. Here, we surveyed 189 human BTB proteins with a cellular fluorescent reporter assay and identified 18 ZBTB TFs that show evidence of polymerization. Through biochemical and cryoelectron microscopy (cryo-EM) studies, we demonstrate that these ZBTB TFs polymerize into filaments. We found that BTB-domain-mediated polymerization of ZBTB TFs enhances chromatin occupancy within regions containing homotypic clusters of TF binding sites, leading to repression of target genes. Our results reveal a role of higher-order structures in regulating ZBTB TFs and suggest an underappreciated role for TF polymerization in modulating gene expression. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44389.map.gz emd_44389.map.gz | 14.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44389-v30.xml emd-44389-v30.xml emd-44389.xml emd-44389.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44389_fsc.xml emd_44389_fsc.xml | 6.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_44389.png emd_44389.png | 64.6 KB | ||
| Filedesc metadata |  emd-44389.cif.gz emd-44389.cif.gz | 6.2 KB | ||
| Others |  emd_44389_half_map_1.map.gz emd_44389_half_map_1.map.gz emd_44389_half_map_2.map.gz emd_44389_half_map_2.map.gz | 26.7 MB 26.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44389 http://ftp.pdbj.org/pub/emdb/structures/EMD-44389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44389 | HTTPS FTP |
-Validation report
| Summary document |  emd_44389_validation.pdf.gz emd_44389_validation.pdf.gz | 671.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44389_full_validation.pdf.gz emd_44389_full_validation.pdf.gz | 670.9 KB | Display | |
| Data in XML |  emd_44389_validation.xml.gz emd_44389_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_44389_validation.cif.gz emd_44389_validation.cif.gz | 17.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44389 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44389 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44389 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44389 | HTTPS FTP |
-Related structure data
| Related structure data |  9b9rMC  9b9vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44389.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44389.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||
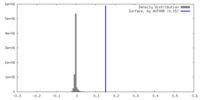
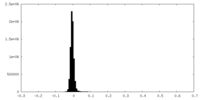
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_44389_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
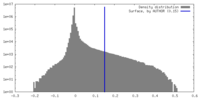
| Density Histograms |
-Half map: #2
| File | emd_44389_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
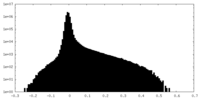
| Density Histograms |
- Sample components
Sample components
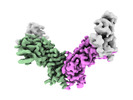
-Entire : ZBTB5 BTB domain filament
| Entire | Name: ZBTB5 BTB domain filament |
|---|---|
| Components |
|
-Supramolecule #1: ZBTB5 BTB domain filament
| Supramolecule | Name: ZBTB5 BTB domain filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Zinc finger and BTB domain-containing protein 5
| Macromolecule | Name: Zinc finger and BTB domain-containing protein 5 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.786326 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDWSHPQFEK SAVGLNDIFE AQKIEWHEGG GGSGENLYFQ GGGRMDFPGH FEQIFQQLNY QRLHGQLCDC VIVVGNRHFK AHRSVLAAC STHFRALFSV AEGDQTMNMI QLDSEVVTAE AFAALIDMMY TSTLMLGESN VMDVLLAASH LHLNSVVKAC K HYLTTRTL UniProtKB: Zinc finger and BTB domain-containing protein 5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1.00 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 50 mM HEPES/NaOH pH 7.4, 200 mM NaCl, 1 mM CHAPSO, 1 mM TCEP | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa Details: Grids were glow-discharged for 60 s at 15-20 mA and 39 Pa. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: LEICA EM GP Details: Grids were vitrified using a Leica EM GP plunge freezer operated at 90% humidity and 10 C with 10 s pre-blot, 3 s blot, 3 s post-blot.. | |||||||||||||||
| Details | Elution fractions from Strep-tag affinity chromatography were dialyzed overnight against 50 mM HEPES/NaOH pH 7.4, 200 mM NaCl, 1 mM TCEP and concentrated by centrifugation. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 10854 / Average exposure time: 2.1 sec. / Average electron dose: 50.7 e/Å2 Details: 2 movies (46 frames) were acquired per hole with 9 holes per stage position. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Details | Real-space refinement without local grid search |
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 194 |
| Output model |  PDB-9b9r: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)