[English] 日本語
 Yorodumi
Yorodumi- EMDB-44266: Cryo-EM structure and molecular assembly of the BoNT-like toxin P... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure and molecular assembly of the BoNT-like toxin PG1 complex from Paeniclostridium ghonii | |||||||||
 Map data Map data | structure and molecular assembly of the BoNT-like toxin PG1 complex from Paeniclostridium ghonii | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PG toxin / TOXIN | |||||||||
| Biological species |  Paraclostridium ghonii (bacteria) Paraclostridium ghonii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Yin L / Dong M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure and molecular assembly of the BoNT-like toxin PG1 complex from Paeniclostridium ghonii Authors: Yin L / Dong M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44266.map.gz emd_44266.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44266-v30.xml emd-44266-v30.xml emd-44266.xml emd-44266.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44266.png emd_44266.png | 56.3 KB | ||
| Masks |  emd_44266_msk_1.map emd_44266_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-44266.cif.gz emd-44266.cif.gz | 6.2 KB | ||
| Others |  emd_44266_half_map_1.map.gz emd_44266_half_map_1.map.gz emd_44266_half_map_2.map.gz emd_44266_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44266 http://ftp.pdbj.org/pub/emdb/structures/EMD-44266 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44266 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44266 | HTTPS FTP |
-Validation report
| Summary document |  emd_44266_validation.pdf.gz emd_44266_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44266_full_validation.pdf.gz emd_44266_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_44266_validation.xml.gz emd_44266_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_44266_validation.cif.gz emd_44266_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44266 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44266 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44266 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44266 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44266.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44266.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | structure and molecular assembly of the BoNT-like toxin PG1 complex from Paeniclostridium ghonii | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
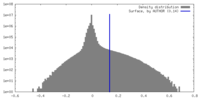
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_44266_msk_1.map emd_44266_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
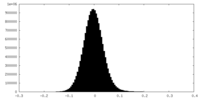
| Density Histograms |
-Half map: half map B
| File | emd_44266_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_44266_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure and molecular assembly of the BoNT-like toxin P...
| Entire | Name: Cryo-EM structure and molecular assembly of the BoNT-like toxin PG1 complex from Paeniclostridium ghonii |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure and molecular assembly of the BoNT-like toxin P...
| Supramolecule | Name: Cryo-EM structure and molecular assembly of the BoNT-like toxin PG1 complex from Paeniclostridium ghonii type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: BoNT-like toxin PG1 complex from Paeniclostridium ghonii |
|---|---|
| Source (natural) | Organism:  Paraclostridium ghonii (bacteria) Paraclostridium ghonii (bacteria) |
-Macromolecule #1: PG1 light chain
| Macromolecule | Name: PG1 light chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Paraclostridium ghonii (bacteria) Paraclostridium ghonii (bacteria) |
| Molecular weight | Theoretical: 44.494844 KDa |
| Recombinant expression | Organism:  Escherichia phage EcSzw-2 (virus) Escherichia phage EcSzw-2 (virus) |
| Sequence | String: MIPINIKDFN YSDPVNNQDI ILVKNEKGSF DKGFFVADKI LLVPARYGNI STDEGGITSK KEKAHVDKKI YLETDSEKNE YLKNMTTLL KRMNSYSTGN KLLNLIIKGE PIYSKDLQGK FIEQTPSRYL DTNTGKRRVN VMITGPGSNV LTKKCTHNGM G LENDPNGK ...String: MIPINIKDFN YSDPVNNQDI ILVKNEKGSF DKGFFVADKI LLVPARYGNI STDEGGITSK KEKAHVDKKI YLETDSEKNE YLKNMTTLL KRMNSYSTGN KLLNLIIKGE PIYSKDLQGK FIEQTPSRYL DTNTGKRRVN VMITGPGSNV LTKKCTHNGM G LENDPNGK HSNGTGILST IEFSPNYLIA YNKCVADPVL TLFHELVHSM HNLYGIAFPD NVKVPYNALK DKNLVSGEEA LS EILTFGG KDLTTEHLET LWKKLAETVI IVKDFVKTDT QAKDVFLNNL RFLSKNENIK IDTIEDIVNG TLKIKNNISN LTE CEFCKE IGDVRIRTRY AVHSEDVTPV EVVDFKNNYK LNSGFLEGQD ISKKYFITNP PKMRRRALRN FKCTIQ |
-Macromolecule #2: PG1 heavy chain
| Macromolecule | Name: PG1 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Paraclostridium ghonii (bacteria) Paraclostridium ghonii (bacteria) |
| Molecular weight | Theoretical: 95.786078 KDa |
| Recombinant expression | Organism:  Escherichia phage EcSzw-2 (virus) Escherichia phage EcSzw-2 (virus) |
| Sequence | String: MADIIASVDK KDVFAVSDTS YFKNFKFPSK KISDTGEVID STKLPQIKDT YKSSREEPIP DNDSTINVKN ITTYHYLEAQ KPKNSSIEL TMVAPSKSKK PNDCVVEAIN DNNKIYTPFS GTAKQFNTVV PIANTAANVI TWLEAIADIF SSETGTFDKL E RAGKETLY ...String: MADIIASVDK KDVFAVSDTS YFKNFKFPSK KISDTGEVID STKLPQIKDT YKSSREEPIP DNDSTINVKN ITTYHYLEAQ KPKNSSIEL TMVAPSKSKK PNDCVVEAIN DNNKIYTPFS GTAKQFNTVV PIANTAANVI TWLEAIADIF SSETGTFDKL E RAGKETLY YIPYVGQLLS IGENVLIGDF KNALLNTGLI ILLDIAPELN IPLLGAFEAY KEYKSLEEFR KAIDNVIDER NK RWHSVYS FVAHQWYGQV NIQIEQRLNH FYQALSYQAG VIKNRVDIEY ARHKEGLEEK EERKLMWASV DCIGSIEASV KEA TKNAEK FLEKSSILYF KEEILPKVHK NLEEFDKNTL FNIYTNIDEF SNRGIAEISE CKKVEADVNN GFRPIKFDFS LLTN LMKSD SLTDEVILEK ALEDALVFSL GVRNGKIQNL SKKWANLTIG TDIRVVHGRD NESIRLNSTQ DSSIQIEKNT NLRFL DSEN FSLSFWIRVP RYNKFDKDKD LNNEYTIVNN MDTATKGFKI SIKNGILLWT LKGTQQKTIE IPLSNTKVSD NIWRHV AII NNKDGNCTIY VDGAQKNAVS LSGLDEITNT LPITLQLVGN KNKKQFIRLD QFNIYEKALS QTEVGKLFSS YFKDSDI RD YWGEPLAYNK TYNMINIAYQ GRGLQSTNNK ISLQPKAVFD PTGDGSYIPR LYRGYDVLLQ KDSQSKTTDI MPKKDDLI N IKLKSGHNFV GFNSTIDTSQ KYLKLTTALL SEVDDPKGFK LMSLKKDNWI QIKKETWMSK NGNVIPQGLV GKRSVDSDV YLYLWDWETE KDDYSEKQWS FICQDEGWID SDGMFTNA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Material: COPPER / Mesh: 400 | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 53.19 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Number classes used: 1 / Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 4.4.1) / Number images used: 364136 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: cryoSPARC (ver. 4.4.1) |
| Final angle assignment | Type: OTHER / Software - Name: cryoSPARC (ver. 4.4.1) |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-9b6m: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)