+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycolicibacterium smegmatis MmpL4 structure | |||||||||
 Map data Map data | Mycolicibacterium smegmatis MmpL4 structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Siderophore / MmpL4 / TRANSPORT PROTEIN | |||||||||
| Function / homology | Membrane transport protein MmpL family / : / Membrane transport protein MMPL domain / MMPL family / plasma membrane / MmpL4 protein Function and homology information Function and homology information | |||||||||
| Biological species |  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) | |||||||||
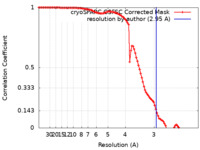
| Method | single particle reconstruction / cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Maharjan R / Klenotic PA / Zhang Z / Yu EW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Mycolicibacterium smegmatis MmpL4 structure Authors: Maharjan R / Klenotic PA / Zhang Z / Yu EW | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44167.map.gz emd_44167.map.gz | 52 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44167-v30.xml emd-44167-v30.xml emd-44167.xml emd-44167.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44167_fsc.xml emd_44167_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_44167.png emd_44167.png | 25.5 KB | ||
| Filedesc metadata |  emd-44167.cif.gz emd-44167.cif.gz | 5.8 KB | ||
| Others |  emd_44167_half_map_1.map.gz emd_44167_half_map_1.map.gz emd_44167_half_map_2.map.gz emd_44167_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44167 http://ftp.pdbj.org/pub/emdb/structures/EMD-44167 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44167 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44167 | HTTPS FTP |
-Validation report
| Summary document |  emd_44167_validation.pdf.gz emd_44167_validation.pdf.gz | 1022.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44167_full_validation.pdf.gz emd_44167_full_validation.pdf.gz | 1022.3 KB | Display | |
| Data in XML |  emd_44167_validation.xml.gz emd_44167_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_44167_validation.cif.gz emd_44167_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44167 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44167 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44167 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44167 | HTTPS FTP |
-Related structure data
| Related structure data |  9b43MC  9b46C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44167.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44167.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mycolicibacterium smegmatis MmpL4 structure | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map A
| File | emd_44167_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_44167_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MmpL4
| Entire | Name: MmpL4 |
|---|---|
| Components |
|
-Supramolecule #1: MmpL4
| Supramolecule | Name: MmpL4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 106.2 kDa/nm |
-Macromolecule #1: MmpL4 protein
| Macromolecule | Name: MmpL4 protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 107.13518 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNPSHDAPT DAMGVSGPKH ARPGGIAKWI RVLAVPIILV WVAIIAVLNT VVPQLDVVGE MRSVSMSPDD APSVIAMKRV GEVFEEFKS NSSVMIVLEG EQPLGDEAHK YYDEIVDKLE ADPAHVEHVQ DFWGDPLTAS GAQSPDGLAS YVQVYTRGNQ G EALANESV ...String: MSNPSHDAPT DAMGVSGPKH ARPGGIAKWI RVLAVPIILV WVAIIAVLNT VVPQLDVVGE MRSVSMSPDD APSVIAMKRV GEVFEEFKS NSSVMIVLEG EQPLGDEAHK YYDEIVDKLE ADPAHVEHVQ DFWGDPLTAS GAQSPDGLAS YVQVYTRGNQ G EALANESV EAVQDIVESV PAPPGVKAYV TGPAALSADQ HVASDRSVRV IEALTFAVII TMLLLVYRSI VTVILTLVMV VL SLSAARG MIAFLGYHEI IGLSVFATNL LTTLAIAAAT DYAIFLIGRY QEARSVGEDR EQSYYTMFHS TAHVVLGSGM TIA GATLCL HFTRMPYFQS LGIPLAIGMS VVVLASLTMG AAIISVASRF GKTFEPKRAM RTRGWRKLGA AVVRWPAPIL VTTI ALSVV GLLALPGYQT NYNDRRYLPQ DLPANTGYAA ADRHFSQARM NPELLMIESD HDLRNSADFL VVDRIAKRVF QVPGI SRVQ AITRPQGTPI EHTSIPFQIS MSGTTQMMNM KYMQDRMADM LVMADEMQKS VDTMEEMLKI TREMSDTTHS MVGKMH GMV EDIKELRDHI ADFDDFLRPI RNYFYWEPHC DSIPVCQSIR SIFDTLDGID VMTDDIQRLM PDMDRLDELM PQMLTIM PP MIESMKTMKT MMLTTQATMG GLQDQMEAAM ENQTAMGQAF DASKNDDSFY LPPETFENPD FKRGMKMFLS PDGHAVRF I ISHEGDPMSP EGIKHIDAIK QAAKEAIKGT PLEGSKIYLG GTAATFKDLQ EGANYDLIIA GIAALCLIFI IMLIITRAV VASAVIVGTV VISLGASFGL SVLIWQHIIG LELHWMVLAM AVIVLLAVGA DYNLLLVSRI KEEIHAGLNT GIIRSMGGTG SVVTSAGLV FAFTMMSMAV SELAVIAQVG TTIGLGLLFD TLVIRSFMTP SIAALMGKWF WWPQRVRQRP LPAPWPQPVQ R DPEDALNR VHHHHHH UniProtKB: MmpL4 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)