[English] 日本語
 Yorodumi
Yorodumi- EMDB-44148: The cryo-EM structure of the H2A.Z-H3.3 double-variant nucleosome -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of the H2A.Z-H3.3 double-variant nucleosome | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | histone variant / nucleosome / chromatin / complex / STRUCTURAL PROTEIN / STRUCTURAL PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of chromosome condensation / Barr body / regulation of centromere complex assembly / pericentric heterochromatin formation / inner kinetochore / muscle cell differentiation / oocyte maturation / nucleus organization / spermatid development / subtelomeric heterochromatin formation ...negative regulation of chromosome condensation / Barr body / regulation of centromere complex assembly / pericentric heterochromatin formation / inner kinetochore / muscle cell differentiation / oocyte maturation / nucleus organization / spermatid development / subtelomeric heterochromatin formation / single fertilization / RNA polymerase II core promoter sequence-specific DNA binding / heterochromatin / Replacement of protamines by nucleosomes in the male pronucleus / nucleosomal DNA binding / Inhibition of DNA recombination at telomere / telomere organization / embryo implantation / RNA Polymerase I Promoter Opening / Assembly of the ORC complex at the origin of replication / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / DNA methylation / Condensation of Prophase Chromosomes / SIRT1 negatively regulates rRNA expression / Chromatin modifications during the maternal to zygotic transition (MZT) / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / cellular response to estradiol stimulus / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / RNA Polymerase I Promoter Escape / Transcriptional regulation by small RNAs / Formation of the beta-catenin:TCF transactivating complex / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / NoRC negatively regulates rRNA expression / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / euchromatin / B-WICH complex positively regulates rRNA expression / multicellular organism growth / heterochromatin formation / Meiotic recombination / Pre-NOTCH Transcription and Translation / cellular response to insulin stimulus / Activation of anterior HOX genes in hindbrain development during early embryogenesis / Transcriptional regulation of granulopoiesis / osteoblast differentiation / male gonad development / structural constituent of chromatin / nucleosome / nucleosome assembly / chromatin organization / RUNX1 regulates transcription of genes involved in differentiation of HSCs / Factors involved in megakaryocyte development and platelet production / Senescence-Associated Secretory Phenotype (SASP) / positive regulation of cell growth / Oxidative Stress Induced Senescence / Estrogen-dependent gene expression / cell population proliferation / chromosome, telomeric region / protein heterodimerization activity / RNA polymerase II cis-regulatory region sequence-specific DNA binding / Amyloid fiber formation / positive regulation of transcription by RNA polymerase II / protein-containing complex / DNA binding / extracellular exosome / extracellular region / nucleoplasm / nucleus Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Tan D / Sokolova V | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Epigenomes / Year: 2024 Journal: Epigenomes / Year: 2024Title: Structural and Biochemical Characterization of the Nucleosome Containing Variants H3.3 and H2A.Z. Authors: Harry Jung / Vladyslava Sokolova / Gahyun Lee / Victoria Rose Stevens / Dongyan Tan /  Abstract: Variant H3.3, along with H2A.Z, is notably enriched at promoter regions and is commonly associated with transcriptional activation. However, the specific molecular mechanisms through which H3.3 ...Variant H3.3, along with H2A.Z, is notably enriched at promoter regions and is commonly associated with transcriptional activation. However, the specific molecular mechanisms through which H3.3 influences chromatin dynamics at transcription start sites, and its role in gene regulation, remain elusive. Using a combination of biochemistry and cryo-electron microscopy (cryo-EM), we show that the inclusion of H3.3 alone does not induce discernible changes in nucleosome DNA dynamics. Conversely, the presence of both H3.3 and H2A.Z enhances DNA's flexibility similarly to H2A.Z alone. Interestingly, our findings suggest that the presence of H3.3 in the H2A.Z nucleosome provides slightly increased protection to DNA at internal sites within the nucleosome. These results imply that while H2A.Z at active promoters promotes the formation of more accessible nucleosomes with increased DNA accessibility to facilitate transcription, the simultaneous presence of H3.3 offers an additional mechanism to fine-tune nucleosome accessibility and the chromatin environment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44148.map.gz emd_44148.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44148-v30.xml emd-44148-v30.xml emd-44148.xml emd-44148.xml | 26.8 KB 26.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44148.png emd_44148.png | 114.1 KB | ||
| Filedesc metadata |  emd-44148.cif.gz emd-44148.cif.gz | 7.2 KB | ||
| Others |  emd_44148_half_map_1.map.gz emd_44148_half_map_1.map.gz emd_44148_half_map_2.map.gz emd_44148_half_map_2.map.gz | 46.2 MB 46.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44148 http://ftp.pdbj.org/pub/emdb/structures/EMD-44148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44148 | HTTPS FTP |
-Validation report
| Summary document |  emd_44148_validation.pdf.gz emd_44148_validation.pdf.gz | 683.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44148_full_validation.pdf.gz emd_44148_full_validation.pdf.gz | 682.9 KB | Display | |
| Data in XML |  emd_44148_validation.xml.gz emd_44148_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_44148_validation.cif.gz emd_44148_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44148 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44148 | HTTPS FTP |
-Related structure data
| Related structure data |  9b3pMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44148.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44148.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
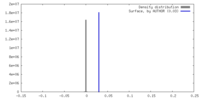
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_44148_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
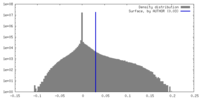
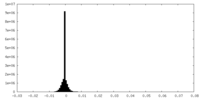
| Density Histograms |
-Half map: #2
| File | emd_44148_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
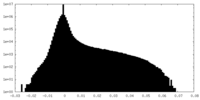
| Density Histograms |
- Sample components
Sample components
-Entire : nucleosome containing histone variants H3.3 and H2A.Z
| Entire | Name: nucleosome containing histone variants H3.3 and H2A.Z |
|---|---|
| Components |
|
-Supramolecule #1: nucleosome containing histone variants H3.3 and H2A.Z
| Supramolecule | Name: nucleosome containing histone variants H3.3 and H2A.Z / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The complex contains Xenopus histone H2B and H4, human H3.3, and mouse H2A.Z |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 288 KDa |
-Macromolecule #1: Histone H3.3
| Macromolecule | Name: Histone H3.3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.360983 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PSTGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSAA IGALQEASEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.3 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 11.394426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A.Z
| Macromolecule | Name: Histone H2A.Z / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.581796 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGGKAGKDS GKAKTKAVSR SQRAGLQFPV GRIHRHLKSR TTSHGRVGAT AAVYSAAILE YLTAEVLELA GNASKDLKVK RITPRHLQL AIRGDEELDS LIKATIAGGG VIPHIHKSLI GKKGQQKTV UniProtKB: Histone H2A.Z |
-Macromolecule #4: Histone H2B 1.1
| Macromolecule | Name: Histone H2B 1.1 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.965265 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPEPAKSAPA PKKGSKKAVT KTQKKDGKKR RKSRKESYAI YVYKVLKQVH PDTGISSKAM SIMNSFVNDV FERIAGEASR LAHYNKRST ITSREIQTAV RLLLPGELAK HAVSEGTKAV TKYTSAK UniProtKB: Histone H2B 1.1 |
-Macromolecule #5: DNA (128-MER)
| Macromolecule | Name: DNA (128-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 45.604047 KDa |
| Sequence | String: (DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT) (DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DG)(DA)(DT) (DT)(DC)(DT) (DC)(DC)(DA)(DG) |
-Macromolecule #6: DNA (128-MER)
| Macromolecule | Name: DNA (128-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 51.269656 KDa |
| Sequence | String: (DA)(DT)(DC)(DC)(DC)(DG)(DC)(DC)(DC)(DT) (DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC)(DC) (DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA)(DT) (DT) (DG)(DG)(DT)(DC)(DG)(DT) ...String: (DA)(DT)(DC)(DC)(DC)(DG)(DC)(DC)(DC)(DT) (DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC)(DC) (DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA)(DT) (DT) (DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA)(DG) (DC)(DA) (DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA)(DC)(DG)(DC)(DA)(DC)(DG)(DT)(DA) (DC)(DG)(DC) (DG)(DC)(DT)(DG)(DT)(DC) (DC)(DC)(DC)(DC)(DG)(DC)(DG)(DT)(DT)(DT) (DT)(DA)(DA)(DC) (DC)(DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG)(DG)(DA)(DT)(DT)(DA)(DC) (DT)(DC)(DC)(DC)(DT) (DA)(DG)(DT)(DC) (DT)(DC)(DC)(DA)(DG)(DG)(DC)(DA)(DC)(DG) (DT)(DG)(DT)(DC)(DA)(DG) (DA)(DT)(DA) (DT)(DA)(DT)(DA)(DC)(DA)(DT)(DC)(DC)(DT) (DG)(DT)(DG)(DC)(DA)(DT)(DG) (DA)(DC) (DT)(DA)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20mM Tris-HCl, 5mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Pressure: 0.0002 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV Details: Freezing condition: blot force 0, blot time 4.5 second. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 5140 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.25 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Details | Phenix real-space refinement was used. | ||||||||||||||||||||||
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 87.42 | ||||||||||||||||||||||
| Output model |  PDB-9b3p: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)