[English] 日本語
 Yorodumi
Yorodumi- EMDB-43985: Tetra-phosphorylated, E1435Q Ycf1 mutant in inward-facing wide co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tetra-phosphorylated, E1435Q Ycf1 mutant in inward-facing wide conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC-transporter / ycf1 / r-domain / phosphorylation / heavy metal / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type Cd2+ transporter / ABC-type cadmium transporter activity / Recycling of bile acids and salts / Heme degradation / Aspirin ADME / Atorvastatin ADME / Paracetamol ADME / P-type cadmium transporter activity / bilirubin transmembrane transporter activity / bilirubin transport ...ABC-type Cd2+ transporter / ABC-type cadmium transporter activity / Recycling of bile acids and salts / Heme degradation / Aspirin ADME / Atorvastatin ADME / Paracetamol ADME / P-type cadmium transporter activity / bilirubin transmembrane transporter activity / bilirubin transport / vacuole fusion, non-autophagic / ABC-family proteins mediated transport / ABC-type glutathione-S-conjugate transporter / ABC-type glutathione S-conjugate transporter activity / fungal-type vacuole membrane / response to metal ion / ATPase-coupled transmembrane transporter activity / response to cadmium ion / glutathione metabolic process / cell redox homeostasis / transmembrane transport / membrane raft / ATP hydrolysis activity / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.23 Å | |||||||||
 Authors Authors | Carvalho RSA / Rasel MSI / Khandelwal NK / Tomasiak TM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of the tetra-phosphorylated R-domain in Ycf1 reveals key interactions for transport regulation Authors: Carvalho RSA / Rasel MSI / Khandelwal NK / Tomasiak TM | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43985.map.gz emd_43985.map.gz | 162.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43985-v30.xml emd-43985-v30.xml emd-43985.xml emd-43985.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43985.png emd_43985.png | 41.8 KB | ||
| Filedesc metadata |  emd-43985.cif.gz emd-43985.cif.gz | 7.3 KB | ||
| Others |  emd_43985_half_map_1.map.gz emd_43985_half_map_1.map.gz emd_43985_half_map_2.map.gz emd_43985_half_map_2.map.gz | 301.9 MB 301.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43985 http://ftp.pdbj.org/pub/emdb/structures/EMD-43985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43985 | HTTPS FTP |
-Validation report
| Summary document |  emd_43985_validation.pdf.gz emd_43985_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43985_full_validation.pdf.gz emd_43985_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_43985_validation.xml.gz emd_43985_validation.xml.gz | 17.1 KB | Display | |
| Data in CIF |  emd_43985_validation.cif.gz emd_43985_validation.cif.gz | 20.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43985 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43985 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43985 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43985 | HTTPS FTP |
-Related structure data
| Related structure data |  9aycMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43985.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43985.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
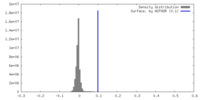
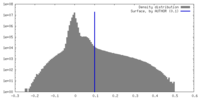
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_43985_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
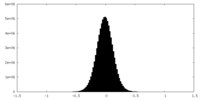
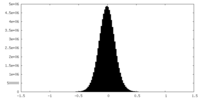
| Density Histograms |
-Half map: #1
| File | emd_43985_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
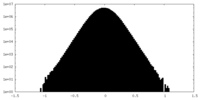
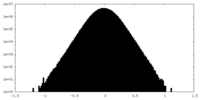
| Density Histograms |
- Sample components
Sample components
-Entire : Phosphorylated, E1435Q Ycf1 mutant in inward-facing wide conformation
| Entire | Name: Phosphorylated, E1435Q Ycf1 mutant in inward-facing wide conformation |
|---|---|
| Components |
|
-Supramolecule #1: Phosphorylated, E1435Q Ycf1 mutant in inward-facing wide conformation
| Supramolecule | Name: Phosphorylated, E1435Q Ycf1 mutant in inward-facing wide conformation type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all / Details: c-AMP PKA treated E1435Q Ycf1 mutant |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 176462 kDa/nm |
-Macromolecule #1: Metal resistance protein YCF1
| Macromolecule | Name: Metal resistance protein YCF1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ABC-type Cd2+ transporter |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 176.263234 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASDYKDDDD KGALEVLFQG PSSPMAGNLV SWACKLCRSP EGFGPISFYG DFTQCFIDGV ILNLSAIFMI TFGIRDLVNL CKKKHSGIK YRRNWIIVSR MALVLLEIAF VSLASLNISK EEAENFTIVS QYASTMLSLF VALALHWIEY DRSVVANTVL L FYWLFETF ...String: MASDYKDDDD KGALEVLFQG PSSPMAGNLV SWACKLCRSP EGFGPISFYG DFTQCFIDGV ILNLSAIFMI TFGIRDLVNL CKKKHSGIK YRRNWIIVSR MALVLLEIAF VSLASLNISK EEAENFTIVS QYASTMLSLF VALALHWIEY DRSVVANTVL L FYWLFETF GNFAKLINIL IRHTYEGIWY SGQTGFILTL FQVITCASIL LLEALPKKPL MPHQHIHQTL TRRKPNPYDS AN IFSRITF SWMSGLMKTG YEKYLVEADL YKLPRNFSSE ELSQKLEKNW ENELKQKSNP SLSWAICRTF GSKMLLAAFF KAI HDVLAF TQPQLLRILI KFVTDYNSER QDDHSSLQGF ENNHPQKLPI VRGFLIAFAM FLVGFTQTSV LHQYFLNVFN TGMY IKSAL TALIYQKSLV LSNEASGLSS TGDIVNLMSV DVQKLQDLTQ WLNLIWSGPF QIIICLYSLY KLLGNSMWVG VIILV IMMP LNSFLMRIQK KLQKSQMKYK DERTRVISEI LNNIKSLKLY AWEKPYREKL EEVRNNKELK NLTKLGCYMA VTSFQF NIV PFLVSCCTFA VFVYTEDRAL TTDLVFPALT LFNLLSFPLM IIPMVLNSFI EASVSIGRLF TFFTNEELQP DSVQRLP KV KNIGDVAINI GDDATFLWQR KPEYKVALKN INFQAKKGNL TCIVGKVGSG KTALLSCMLG DLFRVKGFAT VHGSVAYV S QVPWIMNGTV KENILFGHRY DAEFYEKTIK ACALTIDLAI LMDGDKTLVG EKGISLSGGQ KARLSLARAV YARADTYLL DDPLAAVDEH VARHLIEHVL GPNGLLHTKT KVLATNKVSA LSIADSIALL DNGEITQQGT YDEITKDADS PLWKLLNNYG KKNNGKSNE FGDSSESSVR ESSIPVEGEL EQLQKLNDLD FGNSDAI(SEP)LR RA(SEP)DA(TPO)LG(SEP)I DFGDD ENIA KREHREQGKV KWNIYLEYAK ACNPKSVCVF ILFIVISMFL SVMGNVWLKH WSEVNSRYGS NPNAARYLAI YFALGI GSA LATLIQTIVL WVFCTIHASK YLHNLMTNSV LRAPMTFFET TPIGRILNRF SNDIYKVDAL LGRTFSQFFV NAVKVTF TI TVICATTWQF IFIIIPLSVF YIYYQQYYLR TSRELRRLDS ITRSPIYSHF QETLGGLATV RGYSQQKRFS HINQCRID N NMSAFYPSIN ANRWLAYRLE LIGSIIILGA ATLSVFRLKQ GTLTAGMVGL SLSYALQITQ TLNWIVRMTV EVETNIVSV ERIKEYADLK SEAPLIVEGH RPPKEWPSQG DIKFNNYSTR YRPELDLVLK HINIHIKPNE KVGIVGRTGA GKSSLTLALF RMIEASEGN IVIDNIAINE IGLYDLRHKL SIIPQDSQVF EGTVRENIDP INQYTDEAIW RALELSHLKE HVLSMSNDGL D AQLTEGGG NLSVGQRQLL CLARAMLVPS KILVLDQATA AVDVETDKVV QETIRTAFKD RTILTIAHRL NTIMDSDRII VL DNGKVAE FDSPGQLLSD NKSLFYSLCM EAGLVNENGL VPRGSSAHHH HHHHHH UniProtKB: Metal resistance protein YCF1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.94 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 283 K / Instrument: LEICA EM GP Details: 5 microliters of sample of concentrated E1435Q (5.94 mg/mL) PKA-treated Ycf1 sample was applied to QF-1.2/1,3 Au grid. Grids were place inside of a Leica EM GP2 equilibrated at 10 degress ...Details: 5 microliters of sample of concentrated E1435Q (5.94 mg/mL) PKA-treated Ycf1 sample was applied to QF-1.2/1,3 Au grid. Grids were place inside of a Leica EM GP2 equilibrated at 10 degress Celcius amd 80% humidity. Following 10 seconds incubation, the side of the grid to which the sample was applied was blotted on a Whatman 1 paper (3.5 seconds) then immediately plunge frrozen in liquid ethan equilibrated at -185 degree Celcius.. | ||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 1 / Number real images: 8587 / Average exposure time: 0.9 sec. / Average electron dose: 54.0 e/Å2 Details: Movies were collected at 22,500X magnification with automated super-resolution mode and defocus range of -0.9 to -2.1 micrometer. Moview frames cotained 60 frames with a per frame expousre ...Details: Movies were collected at 22,500X magnification with automated super-resolution mode and defocus range of -0.9 to -2.1 micrometer. Moview frames cotained 60 frames with a per frame expousre of 0.9 electrons per angstrom squared (54 electron/angstrom square total dose) |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)