+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chlamydomonas reinhardtii mastigoneme (constituent map 3) | |||||||||||||||||||||
 Map data Map data | Chlamydomonas reinhardtii mastigoneme (constituent map 3, sharpened) | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Mastigoneme / cilia / STRUCTURAL PROTEIN | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
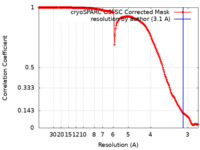
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||||||||
 Authors Authors | Dai J / Ma M / Zhang R / Brown A | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices. Authors: Jin Dai / Meisheng Ma / Qingwei Niu / Robyn J Eisert / Xiangli Wang / Poulomi Das / Karl F Lechtreck / Susan K Dutcher / Rui Zhang / Alan Brown /  Abstract: Hydroxyproline-rich glycoproteins (HRGPs) are a ubiquitous class of protein in the extracellular matrices and cell walls of plants and algae, yet little is known of their native structures or ...Hydroxyproline-rich glycoproteins (HRGPs) are a ubiquitous class of protein in the extracellular matrices and cell walls of plants and algae, yet little is known of their native structures or interactions. Here, we used electron cryomicroscopy (cryo-EM) to determine the structure of the hydroxyproline-rich mastigoneme, an extracellular filament isolated from the cilia of the alga Chlamydomonas reinhardtii. The structure demonstrates that mastigonemes are formed from two HRGPs (a filament of MST1 wrapped around a single copy of MST3) that both have hyperglycosylated poly(hydroxyproline) helices. Within the helices, O-linked glycosylation of the hydroxyproline residues and O-galactosylation of interspersed serine residues create a carbohydrate casing. Analysis of the associated glycans reveals how the pattern of hydroxyproline repetition determines the type and extent of glycosylation. MST3 possesses a PKD2-like transmembrane domain that forms a heteromeric polycystin-like cation channel with PKD2 and SIP, explaining how mastigonemes are tethered to ciliary membranes. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43891.map.gz emd_43891.map.gz | 483.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43891-v30.xml emd-43891-v30.xml emd-43891.xml emd-43891.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43891_fsc.xml emd_43891_fsc.xml | 16.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_43891.png emd_43891.png | 43.4 KB | ||
| Masks |  emd_43891_msk_1.map emd_43891_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43891.cif.gz emd-43891.cif.gz | 4.5 KB | ||
| Others |  emd_43891_additional_1.map.gz emd_43891_additional_1.map.gz emd_43891_half_map_1.map.gz emd_43891_half_map_1.map.gz emd_43891_half_map_2.map.gz emd_43891_half_map_2.map.gz | 257 MB 474.4 MB 474.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43891 http://ftp.pdbj.org/pub/emdb/structures/EMD-43891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43891 | HTTPS FTP |
-Validation report
| Summary document |  emd_43891_validation.pdf.gz emd_43891_validation.pdf.gz | 956.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43891_full_validation.pdf.gz emd_43891_full_validation.pdf.gz | 956.2 KB | Display | |
| Data in XML |  emd_43891_validation.xml.gz emd_43891_validation.xml.gz | 26.3 KB | Display | |
| Data in CIF |  emd_43891_validation.cif.gz emd_43891_validation.cif.gz | 34.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43891 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43891 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43891 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43891 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43891.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43891.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chlamydomonas reinhardtii mastigoneme (constituent map 3, sharpened) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43891_msk_1.map emd_43891_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
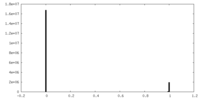
| Density Histograms |
-Additional map: Chlamydomonas reinhardtii mastigoneme (constituent map 3, unsharpened)...
| File | emd_43891_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chlamydomonas reinhardtii mastigoneme (constituent map 3, unsharpened) | ||||||||||||
| Projections & Slices |
| ||||||||||||
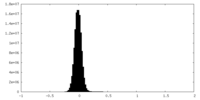
| Density Histograms |
-Half map: Half map A
| File | emd_43891_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
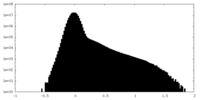
| Density Histograms |
-Half map: Half map B
| File | emd_43891_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
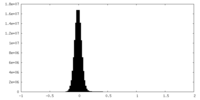
| Density Histograms |
- Sample components
Sample components
-Entire : Native mastigonemes
| Entire | Name: Native mastigonemes |
|---|---|
| Components |
|
-Supramolecule #1: Native mastigonemes
| Supramolecule | Name: Native mastigonemes / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: HMDEKP buffer (30 mM HEPES, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, 25 mM KCl, pH 7.4) containing 1x ProteaseArrest protease inhibitors (G-Biosciences) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number real images: 19601 / Average exposure time: 9.0 sec. / Average electron dose: 38.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)