+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Aca2 from Pectobacterium phage ZF40 bound to RNA | ||||||||||||
 Map data Map data | Sharpened map of Aca2 bound to RNA | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HTH protein / CRISPR / RNA-binding protein / RNA BINDING PROTEIN | ||||||||||||
| Function / homology | Protein of unknown function DUF1870 / Domain of unknown function (DUF1870) / YdiL domain superfamily / Lambda repressor-like, DNA-binding domain superfamily / DNA binding / metal ion binding / DUF1870 family protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Pectobacterium phage ZF40 (virus) Pectobacterium phage ZF40 (virus) | ||||||||||||
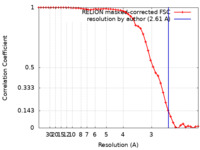
| Method | single particle reconstruction / cryo EM / Resolution: 2.61 Å | ||||||||||||
 Authors Authors | Wilkinson ME / Birkholz N / Kimanius D / Fineran PC | ||||||||||||
| Funding support |  United States, United States,  New Zealand, New Zealand,  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Phage anti-CRISPR control by an RNA- and DNA-binding helix-turn-helix protein. Authors: Nils Birkholz / Kotaro Kamata / Maximilian Feussner / Max E Wilkinson / Christian Cuba Samaniego / Angela Migur / Dari Kimanius / Marijn Ceelen / Sam C Went / Ben Usher / Tim R Blower / ...Authors: Nils Birkholz / Kotaro Kamata / Maximilian Feussner / Max E Wilkinson / Christian Cuba Samaniego / Angela Migur / Dari Kimanius / Marijn Ceelen / Sam C Went / Ben Usher / Tim R Blower / Chris M Brown / Chase L Beisel / Zasha Weinberg / Robert D Fagerlund / Simon A Jackson / Peter C Fineran /      Abstract: In all organisms, regulation of gene expression must be adjusted to meet cellular requirements and frequently involves helix-turn-helix (HTH) domain proteins. For instance, in the arms race between ...In all organisms, regulation of gene expression must be adjusted to meet cellular requirements and frequently involves helix-turn-helix (HTH) domain proteins. For instance, in the arms race between bacteria and bacteriophages, rapid expression of phage anti-CRISPR (acr) genes upon infection enables evasion from CRISPR-Cas defence; transcription is then repressed by an HTH-domain-containing anti-CRISPR-associated (Aca) protein, probably to reduce fitness costs from excessive expression. However, how a single HTH regulator adjusts anti-CRISPR production to cope with increasing phage genome copies and accumulating acr mRNA is unknown. Here we show that the HTH domain of the regulator Aca2, in addition to repressing Acr synthesis transcriptionally through DNA binding, inhibits translation of mRNAs by binding conserved RNA stem-loops and blocking ribosome access. The cryo-electron microscopy structure of the approximately 40 kDa Aca2-RNA complex demonstrates how the versatile HTH domain specifically discriminates RNA from DNA binding sites. These combined regulatory modes are widespread in the Aca2 family and facilitate CRISPR-Cas inhibition in the face of rapid phage DNA replication without toxic acr overexpression. Given the ubiquity of HTH-domain-containing proteins, it is anticipated that many more of them elicit regulatory control by dual DNA and RNA binding. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43762.map.gz emd_43762.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43762-v30.xml emd-43762-v30.xml emd-43762.xml emd-43762.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43762_fsc.xml emd_43762_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_43762.png emd_43762.png | 122.6 KB | ||
| Masks |  emd_43762_msk_1.map emd_43762_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43762.cif.gz emd-43762.cif.gz | 6.3 KB | ||
| Others |  emd_43762_half_map_1.map.gz emd_43762_half_map_1.map.gz emd_43762_half_map_2.map.gz emd_43762_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43762 http://ftp.pdbj.org/pub/emdb/structures/EMD-43762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43762 | HTTPS FTP |
-Validation report
| Summary document |  emd_43762_validation.pdf.gz emd_43762_validation.pdf.gz | 810.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43762_full_validation.pdf.gz emd_43762_full_validation.pdf.gz | 810.3 KB | Display | |
| Data in XML |  emd_43762_validation.xml.gz emd_43762_validation.xml.gz | 9.9 KB | Display | |
| Data in CIF |  emd_43762_validation.cif.gz emd_43762_validation.cif.gz | 13.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43762 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43762 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43762 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43762 | HTTPS FTP |
-Related structure data
| Related structure data |  8w35MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43762.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43762.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of Aca2 bound to RNA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0618 Å | ||||||||||||||||||||||||||||||||||||
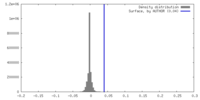
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43762_msk_1.map emd_43762_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
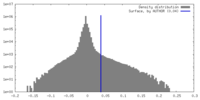
| Density Histograms |
-Half map: Aca2-RNA refinement, half-map 1
| File | emd_43762_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Aca2-RNA refinement, half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
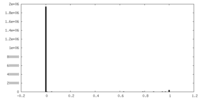
| Density Histograms |
-Half map: Aca2-RNA refinement, half-map 2
| File | emd_43762_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Aca2-RNA refinement, half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
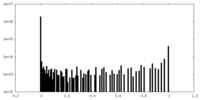
| Density Histograms |
- Sample components
Sample components
-Entire : Anti-CRISPR associated (Aca) protein, Aca2 bound to its 5'UTR RNA...
| Entire | Name: Anti-CRISPR associated (Aca) protein, Aca2 bound to its 5'UTR RNA (IR2 and IR-RBS) |
|---|---|
| Components |
|
-Supramolecule #1: Anti-CRISPR associated (Aca) protein, Aca2 bound to its 5'UTR RNA...
| Supramolecule | Name: Anti-CRISPR associated (Aca) protein, Aca2 bound to its 5'UTR RNA (IR2 and IR-RBS) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pectobacterium phage ZF40 (virus) Pectobacterium phage ZF40 (virus) |
| Molecular weight | Theoretical: 39.5 KDa |
-Macromolecule #1: Anti-CRISPR associated (Aca) protein, Aca2
| Macromolecule | Name: Anti-CRISPR associated (Aca) protein, Aca2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pectobacterium phage ZF40 (virus) Pectobacterium phage ZF40 (virus) |
| Molecular weight | Theoretical: 13.710401 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SHGSMTNKEL QAIRKLLMLD VSEAAEHIGR VSARSWQYWE SGRSAVPDDV EQEMLDLASV RIEMMSAIDK RLADGERPKL RFYNKLDEY LADNPDHNVI GWRLSQSVAA LYYTEGHADL I UniProtKB: DUF1870 family protein |
-Macromolecule #2: IR2 and IR-RBS RNA
| Macromolecule | Name: IR2 and IR-RBS RNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Pectobacterium phage ZF40 (virus) Pectobacterium phage ZF40 (virus) |
| Molecular weight | Theoretical: 13.463021 KDa |
| Sequence | String: AUCGGUUCGA GAUGGCUCGA AUCGCUCCUA ACGAGGAUUC CA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 25 mA plasma | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 285 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 11232 / Average exposure time: 1.33 sec. / Average electron dose: 72.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8w35: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)