+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | HTT in complex with HAP40 in the apo state. | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Huntington's Disease / neurodegenerative disease / UNKNOWN FUNCTION | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報vesicle cytoskeletal trafficking / regulation of cAMP-dependent protein kinase activity / regulation of phosphoprotein phosphatase activity / positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity / microtubule-based transport / vocal learning / regulation of CAMKK-AMPK signaling cascade / negative regulation of proteasomal protein catabolic process / positive regulation of mitophagy / profilin binding ...vesicle cytoskeletal trafficking / regulation of cAMP-dependent protein kinase activity / regulation of phosphoprotein phosphatase activity / positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity / microtubule-based transport / vocal learning / regulation of CAMKK-AMPK signaling cascade / negative regulation of proteasomal protein catabolic process / positive regulation of mitophagy / profilin binding / vesicle transport along microtubule / positive regulation of cilium assembly / presynaptic cytosol / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / positive regulation of aggrephagy / postsynaptic cytosol / positive regulation of lipophagy / dynein intermediate chain binding / beta-tubulin binding / Golgi organization / establishment of mitotic spindle orientation / dynactin binding / Regulation of MECP2 expression and activity / autophagosome / inclusion body / heat shock protein binding / centriole / negative regulation of extrinsic apoptotic signaling pathway / protein destabilization / cytoplasmic vesicle membrane / kinase binding / p53 binding / late endosome / transmembrane transporter binding / early endosome / nuclear body / positive regulation of apoptotic process / axon / apoptotic process / dendrite / perinuclear region of cytoplasm / Golgi apparatus / endoplasmic reticulum / protein-containing complex / nucleoplasm / identical protein binding / nucleus / cytoplasm / cytosol 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.72 Å | |||||||||
 データ登録者 データ登録者 | Poweleit N / Boudet J / Doherty E | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: To Be Published ジャーナル: To Be Publishedタイトル: Discovery of a Small Molecule Ligand to the Huntingtin/HAP40 complex 著者: Poweleit N / Boudet J / Doherty E | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
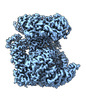
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_43715.map.gz emd_43715.map.gz | 91.6 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-43715-v30.xml emd-43715-v30.xml emd-43715.xml emd-43715.xml | 19.6 KB 19.6 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_43715.png emd_43715.png | 146.5 KB | ||
| Filedesc metadata |  emd-43715.cif.gz emd-43715.cif.gz | 7.9 KB | ||
| その他 |  emd_43715_half_map_1.map.gz emd_43715_half_map_1.map.gz emd_43715_half_map_2.map.gz emd_43715_half_map_2.map.gz | 107.3 MB 107.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43715 http://ftp.pdbj.org/pub/emdb/structures/EMD-43715 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43715 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43715 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_43715_validation.pdf.gz emd_43715_validation.pdf.gz | 777.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_43715_full_validation.pdf.gz emd_43715_full_validation.pdf.gz | 776.9 KB | 表示 | |
| XML形式データ |  emd_43715_validation.xml.gz emd_43715_validation.xml.gz | 14 KB | 表示 | |
| CIF形式データ |  emd_43715_validation.cif.gz emd_43715_validation.cif.gz | 16.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43715 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43715 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43715 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43715 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8w15MC  8vlxC C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_43715.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_43715.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ボクセルのサイズ | X=Y=Z: 0.83 Å | ||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
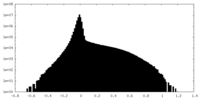
-ハーフマップ: #2
| ファイル | emd_43715_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
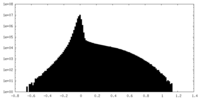
-ハーフマップ: #1
| ファイル | emd_43715_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : HTT and HAP40 complex
| 全体 | 名称: HTT and HAP40 complex |
|---|---|
| 要素 |
|
-超分子 #1: HTT and HAP40 complex
| 超分子 | 名称: HTT and HAP40 complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Huntingtin
| 分子 | 名称: Huntingtin / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 353.493656 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MATLEKLMKA FESLKSFQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQPPPPP PPPPPPQLPQ PPPQAQPLL PQPQPPPPPP PPPPGPAVAE EPLHRPKKEL SATKKDRVNH CLTICENIVA QSVRNSPEFQ KLLGIAMELF L LCSDDAES ...文字列: MATLEKLMKA FESLKSFQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQPPPPP PPPPPPQLPQ PPPQAQPLL PQPQPPPPPP PPPPGPAVAE EPLHRPKKEL SATKKDRVNH CLTICENIVA QSVRNSPEFQ KLLGIAMELF L LCSDDAES DVRMVADECL NKVIKALMDS NLPRLQLELY KEIKKNGAPR SLRAALWRFA ELAHLVRPQK CRPYLVNLLP CL TRTSKRP EESVQETLAA AVPKIMASFG NFANDNEIKV LLKAFIANLK SSSPTIRRTA AGSAVSICQH SRRTQYFYSW LLN VLLGLL VPVEDEHSTL LILGVLLTLR YLVPLLQQQV KDTSLKGSFG VTRKEMEVSP SAEQLVQVYE LTLHHTQHQD HNVV TGALE LLQQLFRTPP PELLQTLTAV GGIGQLTAAK EESGGRSRSG SIVELIAGGG SSCSPVLSRK QKGKVLLGEE EALED DSES RSDVSSSALT ASVKDEISGE LAASSGVSTP GSAGHDIITE QPRSQHTLQA DSVDLASCDL TSSATDGDEE DILSHS SSQ VSAVPSDPAM DLNDGTQASS PISDSSQTTT EGPDSAVTPS DSSEIVLDGT DNQYLGLQIG QPQDEDEEAT GILPDEA SE AFRNSSMALQ QAHLLKNMSH CRQPSDSSVD KFVLRDEATE PGDQENKPCR IKGDIGQSTD DDSAPLVHCV RLLSASFL L TGGKNVLVPD RDVRVSVKAL ALSCVGAAVA LHPESFFSKL YKVPLDTTEY PEEQYVSDIL NYIDHGDPQV RGATAILCG TLICSILSRS RFHVGDWMGT IRTLTGNTFS LADCIPLLRK TLKDESSVTC KLACTAVRNC VMSLCSSSYS ELGLQLIIDV LTLRNSSYW LVRTELLETL AEIDFRLVSF LEAKAENLHR GAHHYTGLLK LQERVLNNVV IHLLGDEDPR VRHVAAASLI R LVPKLFYK CDQGQADPVV AVARDQSSVY LKLLMHETQP PSHFSVSTIT RIYRGYNLLP SITDVTMENN LSRVIAAVSH EL ITSTTRA LTFGCCEALC LLSTAFPVCI WSLGWHCGVP PLSASDESRK SCTVGMATMI LTLLSSAWFP LDLSAHQDAL ILA GNLLAA SAPKSLRSSW ASEEEANPAA TKQEEVWPAL GDRALVPMVE QLFSHLLKVI NICAHVLDDV APGPAIKAAL PSLT NPPSL SPIRRKGKEK EPGEQASVPL SPKKGSEASA ASRQSDTSGP VTTSKSSSLG SFYHLPSYLK LHDVLKATHA NYKVT LDLQ NSTEKFGGFL RSALDVLSQI LELATLQDIG KCVEEILGYL KSCFSREPMM ATVCVQQLLK TLFGTNLASQ FDGLSS NPS KSQGRAQRLG SSSVRPGLYH YCFMAPYTHF TQALADASLR NMVQAEQEND TSGWFDVLQK VSTQLKTNLT SVTKNRA DK NAIHNHIRLF EPLVIKALKQ YTTTTCVQLQ KQVLDLLAQL VQLRVNYCLL DSDQVFIGFV LKQFEYIEVG QFRESEAI I PNIFFFLVLL SYERYHSKQI IGIPKIIQLC DGIMASGRKA VTHAIPALQP IVHDLFVLRG TNKADAGKEL ETQKEVVVS MLLRLIQYHQ VLEMFILVLQ QCHKENEDKW KRLSRQIADI ILPMLAKQQM HIDSHEALGV LNTLFEILAP SSLRPVDMLL RSMFVTPNT MASVSTVQLW ISGILAILRV LISQSTEDIV LSRIQELSFS PYLISCTVIN RLRDGDSTST LEEHSEGKQI K NLPEETFS RFLLQLVGIL LEDIVTKQLK VEMSEQQHTF YCQELGTLLM CLIHIFKSGM FRRITAAATR LFRSDGCGGS FY TLDSLNL RARSMITTHP ALVLLWCQIL LLVNHTDYRW WAEVQQTPKR HSLSSTKLLS PQMSGEEEDS DLAAKLGMCN REI VRRGAL ILFCDYVCQN LHDSEHLTWL IVNHIQDLIS LSHEPPVQDF ISAVHRNSAA SGLFIQAIQS RCENLSTPTM LKKT LQCLE GIHLSQSGAV LTLYVDRLLC TPFRVLARMV DILACRRVEM LLAANLQSSM AQLPMEELNR IQEYLQSSGL AQRHQ RLYS LLDRFRLSTM QDSLSPSPPV SSHPLDGDGH VSLETVSPDK DWYVHLVKSQ CWTRSDSALL EGAELVNRIP AEDMNA FMM NSEFNLSLLA PCLSLGMSEI SGGQKSALFE AAREVTLARV SGTVQQLPAV HHVFQPELPA EPAAYWSKLN DLFGDAA LY QSLPTLARAL AQYLVVVSKL PSHLHLPPEK EKDIVKFVVA TLEALSWHLI HEQIPLSLDL QAGLDCCCLA LQLPGLWS V VSSTEFVTHA CSLIYCVHFI LEAVAVQPGE QLLSPERRTN TPKAISEEEE EVDPNTQNPK YITAACEMVA EMVESLQSV LALGHKRNSG VPAFLTPLLR NIIISLARLP LVNSYTRVPP LVWKLGWSPK PGGDFGTAFP EIPVEFLQEK EVFKEFIYRI NTLGWTSRT QFEETWATLL GVLVTQPLVM EQEESPPEED TERTQINVLA VQAITSLVLS AMTVPVAGNP AVSCLEQQPR N KPLKALDT RFGRKLSIIR GIVEQEIQAM VSKRENIATH HLYQAWDPVP SLSPATTGAL ISHEKLLLQI NPERELGSMS YK LGQVSIH SVWLGNSITP LREEEWDEEE EEEADAPAPS SPPTSPVNSR KHRAGVDIHS CSQFLLELYS RWILPSSSAR RTP AILISE VVRSLLVVSD LFTERNQFEL MYVTLTELRR VHPSEDEILA QYLVPATCKA AAVLGMDKAV AEPVSRLLES TLRS SHLPS RVGALHGVLY VLECDLLDDT AKQLIPVISD YLLSNLKGIA HCVNIHSQQH VLVMCATAFY LIENYPLDVG PEFSA SIIQ MCGVMLSGSE ESTPSIIYHC ALRGLERLLL SEQLSRLDAE SLVKLSVDRV NVHSPHRAMA ALGLMLTCMY TGKEKV SPG RTSDPNPAAP DSESVIVAME RVSVLFDRIR KGFPCEARVV ARILPQFLDD FFPPQDIMNK VIGEFLSNQQ PYPQFMA TV VYKVFQTLHS TGQSSMVRDW VMLSLSNFTQ RAPVAMATWS LSCFFVSAST SPWVAAILPH VISRMGKLEQ VDVNLFCL V ATDFYRHQIE EELDRRAFQS VLEVVAAPGS PYHRLLTCLR NVHKVTTCAA AENLYFQGDY KDDDDK UniProtKB: Huntingtin |
-分子 #2: 40-kDa huntingtin-associated protein
| 分子 | 名称: 40-kDa huntingtin-associated protein / タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 41.342254 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MHHHHHHSSG RENLYFQGMA AAAAGLGGGG AGPGPEAGDF LARYRLVSNK LKKRFLRKPN VAEAGEQFGQ LGRELRAQEC LPYAAWCQL AVARCQQALF HGPGEALALT EAARLFLRQE RDARQRLVCP AAYGEPLQAA ASALGAAVRL HLELGQPAAA A ALCLELAA ...文字列: MHHHHHHSSG RENLYFQGMA AAAAGLGGGG AGPGPEAGDF LARYRLVSNK LKKRFLRKPN VAEAGEQFGQ LGRELRAQEC LPYAAWCQL AVARCQQALF HGPGEALALT EAARLFLRQE RDARQRLVCP AAYGEPLQAA ASALGAAVRL HLELGQPAAA A ALCLELAA ALRDLGQPAA AAGHFQRAAQ LQLPQLPLAA LQALGEAASC QLLARDYTGA LAVFTRMQRL AREHGSHPVQ SL PPPPPPA PQPGPGATPA LPAALLPPNS GSAAPSPAAL GAFSDVLVRC EVSRVLLLLL LQPPPAKLLP EHAQTLEKYS WEA FDSHGQ ESSGQLPEEL FLLLQSLVMA THEKDTEAIK SLQVEMWPLL TAEQNHLLHL VLQETISPSG QGV UniProtKB: 40-kDa huntingtin-associated protein |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 詳細: 25 mM Hepes, 300 mM NaCl, 0.025%CHAPS, 1mM DTT, 1% DMSO |
|---|---|
| グリッド | モデル: Quantifoil R2/4 / 支持フィルム - 材質: GRAPHENE OXIDE / 支持フィルム - トポロジー: CONTINUOUS |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 4 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 撮影したグリッド数: 1 / 実像数: 6788 / 平均露光時間: 1.4 sec. / 平均電子線量: 50.48 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.0 µm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT |
| 得られたモデル |  PDB-8w15: |
 ムービー
ムービー コントローラー
コントローラー








 Z
Z Y
Y X
X